Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle
- PMID: 9696870
- PMCID: PMC110032
- DOI: 10.1128/JVI.72.9.7653-7658.1998
Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle
Abstract
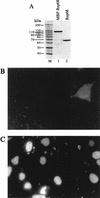
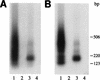
Adeno-associated virus (AAV) integrates very efficiently into a specific site (AAVS1) of human chromosome 19. Two elements of the AAV genome are sufficient: the inverted terminal repeats (ITRs) and the Rep78 or Rep68 protein. The incorporation of the AAV integration machinery in nonviral delivery systems is of great interest for gene therapy. We demonstrate that purified recombinant Rep68 protein is functionally active when directly delivered into human cells by using the polycationic liposome Lipofectamine, promoting the rescue-replication of a codelivered ITR-flanked cassette in adenovirus-infected cells and its site-specific integration in noninfected cells. The sequencing of cloned virus-host DNA junctions confirmed that lipofected Rep68 protein triggers site-specific integration at the same sites in chromosome 19 already characterized in cells latently infected with AAV.
Figures




Similar articles
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19.J Virol. 2002 Jul;76(14):7163-73. doi: 10.1128/jvi.76.14.7163-7173.2002. J Virol. 2002. PMID: 12072516 Free PMC article.
-
A Rep recognition sequence is necessary but not sufficient for nicking of DNA by adeno-associated virus type-2 Rep proteins.Arch Biochem Biophys. 2001 May 15;389(2):271-7. doi: 10.1006/abbi.2001.2348. Arch Biochem Biophys. 2001. PMID: 11339817
-
Preferential integration of adeno-associated virus type 2 into a polypyrimidine/polypurine-rich region within AAVS1.J Virol. 2007 Sep;81(18):9718-26. doi: 10.1128/JVI.00746-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626070 Free PMC article.
-
Positive and negative effects of adeno-associated virus Rep on AAVS1-targeted integration.J Gen Virol. 2003 Aug;84(Pt 8):2127-2132. doi: 10.1099/vir.0.19193-0. J Gen Virol. 2003. PMID: 12867644
-
Site-specific integration by adeno-associated virus.Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11288-94. doi: 10.1073/pnas.93.21.11288. Proc Natl Acad Sci U S A. 1996. PMID: 8876128 Free PMC article. Review.
Cited by
-
Charge-to-alanine mutagenesis of the adeno-associated virus type 2 Rep78/68 proteins yields temperature-sensitive and magnesium-dependent variants.J Virol. 1999 Nov;73(11):9433-45. doi: 10.1128/JVI.73.11.9433-9445.1999. J Virol. 1999. PMID: 10516052 Free PMC article.
-
Site-specific integration of an adeno-associated virus vector plasmid mediated by regulated expression of rep based on Cre-loxP recombination.J Virol. 2000 Nov;74(22):10631-8. doi: 10.1128/jvi.74.22.10631-10638.2000. J Virol. 2000. PMID: 11044107 Free PMC article.
-
Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment.J Virol. 2000 Aug;74(16):7671-7. doi: 10.1128/jvi.74.16.7671-7677.2000. J Virol. 2000. PMID: 10906224 Free PMC article.
-
Effects of Cellular Methylation on Transgene Expression and Site-Specific Integration of Adeno-Associated Virus.Genes (Basel). 2017 Sep 18;8(9):232. doi: 10.3390/genes8090232. Genes (Basel). 2017. PMID: 28926997 Free PMC article.
-
A chimeric protein containing the N terminus of the adeno-associated virus Rep protein recognizes its target site in an in vivo assay.J Virol. 2000 Mar;74(5):2372-82. doi: 10.1128/jvi.74.5.2372-2382.2000. J Virol. 2000. PMID: 10666268 Free PMC article.
References
-
- Berns K I, Linden R M. The cryptic life style of adeno-associated virus. Bioessays. 1995;17:237–245. - PubMed
-
- Deshnukh H M, Huang L. Liposome and polylysine mediated gene transfer. New J Chem. 1997;21:113–124.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

