Methylation status of CpG sites and methyl-CpG binding proteins are involved in the promoter regulation of the mouse Xist gene
- PMID: 9699479
- PMCID: PMC6190200
Methylation status of CpG sites and methyl-CpG binding proteins are involved in the promoter regulation of the mouse Xist gene
Abstract
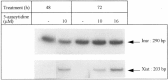
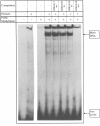
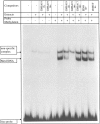
The mouse Xist gene is expressed exclusively from the inactive X chromosome and is involved in the initiation of X inactivation. We previously reported that the -1157/+917 region of the Xist promoter was ubiquitously functional in mammalian cells and that experiments in a transient expression system revealed no trans-acting element responsible for the inactive X specific expression of Xist. In somatic tissues, the 5' end of the silent Xist allele on the active X is known to be fully methylated whereas the expressed allele on the inactive X is unmethylated. In the present study we have used a bisulphite genomic sequencing method to evaluate DNA methylation at all cytosines including CpG dinucleotides within the Xist promoter. We report and confirm that methylation of specific sites plays a key role in Xist gene expression. In vitro DNA methylation of the 5'-region drastically reduced transcriptional activity in transiently transfected fibroblasts. Mobility shift assays showed that methylation does not inhibit Xist promoter activity by preventing the binding of transcription factors and that two distinct nuclear proteins bind in a sequence methyl-CpG-specific manner. Therefore, we suggest that Xist repression involves its promoter methylation and two distinct methylated DNA binding proteins.
Figures



References
-
- Beard C.; Li E.; Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 9:2325–2334; 1995. - PubMed
-
- Borsani G.; Tonlorenzi R.; Simmler M. C.; Dandolo L.; Arnaud D.; Capra V.; Grompe M.; Pizzuti A.; Muzni D.; Lawrence C.; Willard H. F.; Avner P.; Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351:325–329; 1991. - PubMed
-
- Boyes J.; Bird A. DNA methylation inhibits transcription indirectly via methyl-CpG binding protein. Cell 64:1123; 1991. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
