Overexpression of a neural-specific rho family GTPase, cRac1B, selectively induces enhanced neuritogenesis and neurite branching in primary neurons
- PMID: 9700168
- PMCID: PMC2148164
- DOI: 10.1083/jcb.142.3.815
Overexpression of a neural-specific rho family GTPase, cRac1B, selectively induces enhanced neuritogenesis and neurite branching in primary neurons
Abstract
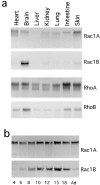
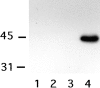
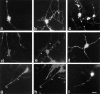
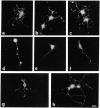
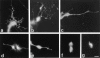
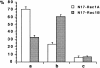
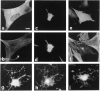
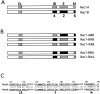
Rho family GTPases have been implicated in cytoskeletal reorganization during neuritogenesis. We have recently identified a new gene of this family, cRac1B, specifically expressed in the chicken developing nervous system. This GTPase was overexpressed in primary neurons to study the role of cRac1B in the development of the neuronal phenotype. Overexpression of cRac1B induced an increment in the number of neurites per neuron, and dramatically increased neurite branching, whereas overexpression of the highly related and ubiquitous cRac1A GTPase did not evidently affect neuronal morphology. Furthermore, expression of an inactive form of cRac1B strikingly inhibited neurite formation. The specificity of cRac1B action observed in neurons was not observed in fibroblasts, where both GTPases produced similar effects on cell morphology and actin organization, indicating the existence of a cell type-dependent specificity of cRac1B function. Molecular dissection of cRac1B function by analysis of the effects of chimeric cRac1A/cRac1B proteins showed that the COOH-terminal portion of cRac1B is essential to induce increased neuritogenesis and neurite branching. Considering the distinctive regulation of cRac1B expression during neural development, our data strongly support an important role of cRac1B during neuritogenesis, and they uncover new mechanisms underlying the functional specificity of distinct Rho family GTPases.
Figures


References
-
- Acheson DWK, Kemplay SK, Webster KE. Quantitative analysis of optic terminal profile distribution within the pigeon optic tectum. Neuroscience. 1980;5:1067–1084. - PubMed
-
- Belloc F, Dumain P, Boisseau MR, Jalloustre C, Reiffers J, Bernard P, Lacombe F. A flow cytometric method using Hoechst 33342 and propidium iodide for simultaneous cell cycle analysis and apoptosis determination in unfixed cells. Cytometry. 1994;17:59–65. - PubMed
-
- Cattelino A, Longhi R, de Curtis I. Differential distribution of two cytoplasmic variants of the α6β1 integrin laminin receptor in the ventral plasma membrane of embryonic fibroblasts. J Cell Sci. 1995;108:3067–3078. - PubMed
-
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

