Characterization of Euphorbia characias latex amine oxidase
- PMID: 9701592
- PMCID: PMC34900
- DOI: 10.1104/pp.117.4.1363
Characterization of Euphorbia characias latex amine oxidase
Abstract
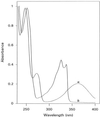
A copper-containing amine oxidase from the latex of Euphorbia characias was purified to homogeneity and the copper-free enzyme obtained by a ligand-exchange procedure. The interactions of highly purified apo- and holoenzyme with several substrates, carbonyl reagents, and copper ligands were investigated by optical spectroscopy under both aerobic and anaerobic conditions. The extinction coefficients at 278 and 490 nm were determined as 3.78 x 10(5) M-1 cm-1 and 6000 M-1 cm-1, respectively. Active-site titration of highly purified enzyme with substrates and carbonyl reagents showed the presence of one cofactor at each enzyme subunit. In anaerobiosis the native enzyme oxidized one equivalent substrate and released one equivalent aldehyde per enzyme subunit. The apoenzyme gave exactly the same 1:1:1 stoichiometry in anaerobiosis and in aerobiosis. These findings demonstrate unequivocally that copper-free amine oxidase can oxidize substrates with a single half-catalytic cycle. The DNA-derived protein sequence shows a characteristic hexapeptide present in most 6-hydroxydopa quinone-containing amine oxidases. This hexapeptide contains the tyrosinyl residue that can be modified into the cofactor 6-hydroxydopa quinone.
Figures






References
-
- Bellelli A, Finazzi Agró A, Floris G, Brunori M. On the mechanism and rate of substrate oxidation by amine oxidase from lentil seedlings. J Biol Chem. 1991;266:20654–20657. - PubMed
-
- Brown DE, McGuirl MA, Dooley DM, Janes SM, Mu D, Klinman JP. The organic functional group in copper-containing amine oxidases. J Biol Chem. 1991;266:4049–4051. - PubMed
-
- Cai D, Klinman JP. Evidence for a self-catalytic mechanism of 2,4,5-trihydroxyphenylalanine quinone biogenesis in yeast copper amine oxidase. J Biol Chem. 1994;269:32039–32042. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

