Species identification and virulence attributes of Saccharomyces boulardii (nom. inval.)
- PMID: 9705402
- PMCID: PMC105172
- DOI: 10.1128/JCM.36.9.2613-2617.1998
Species identification and virulence attributes of Saccharomyces boulardii (nom. inval.)
Abstract
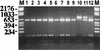
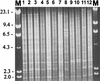
Saccharomyces boulardii (nom. inval.) has been used for the treatment of several types of diarrhea. Recent studies have confirmed that S. boulardii is effective in the treatment of diarrhea, in particular chronic or recurrent diarrhea, and furthermore that it is a safe and well-tolerated treatment. The aim of the present study was to identify strains of S. boulardii to the species level and assess their virulence in established murine models. Three strains of S. boulardii were obtained from commercially available products in France and Italy. The three S. boulardii strains did not form spores upon repeated testing. Therefore, classical methods used for the identification of Saccharomyces spp. could not be undertaken. Typing by using the restriction fragment length polymorphisms (RFLPs) of the PCR-amplified intergenic transcribed spacer regions (including the 5. 8S ribosomal DNA) showed that the three isolates of S. boulardii were not separable from authentic isolates of Saccharomyces cerevisiae with any of the 10 restriction endonucleases assessed, whereas 9 of the 10 recognized species of Saccharomyces could be differentiated. RFLP analysis of cellular DNA with EcoRI showed that all three strains of S. boulardii had identical patterns and were similar to other authentic S. cerevisiae isolates tested. Therefore, the commercial strains of S. boulardii available to us cannot be genotypically distinguished from S. cerevisiae. Two S. boulardii strains were tested in CD-1 and DBA/2N mouse models of systemic disease and showed intermediate virulence compared with virulent and avirulent strains of S. cerevisiae. The results of the present study show that these S. boulardii strains are asporogenous strains of the species S. cerevisiae, not representatives of a distinct and separate species, and possess moderate virulence in murine models of systemic infection. Therefore, caution should be advised in the clinical use of these strains in immunocompromised patients until further study is undertaken.
Figures

References
-
- Baleiras Couto M M, Vogels J T, Hofstra H, Huis in’t Veld J H, Vossen J M. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for food-borne yeasts. J Appl Bacteriol. 1995;79:525–535. - PubMed
-
- Banno I, Kaneko Y. A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast. 1989;5:S373–S377. - PubMed
-
- Barnett J A. The taxonomy of the genus Saccharomyces Meyen ex Rees: a short review for the non-taxonomists. Yeast. 1992;8:1–23. - PubMed
-
- Barnett J A, Payne R W, Yarrow D. Yeast characteristics and identification. Cambridge, England: Cambridge University Press; 1990.
-
- Boekhout T, Kurtzman C P, O’Donnell K, Smith M T. Phylogeny of the yeast genera Hanseniaspora (anamorph Kloeckera), Dekkera (anamorph Brettanomyces), and Eeniella as inferred from partial 26S ribosomal DNA nucleotide sequences. Int J Syst Bacteriol. 1994;44:781–786. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

