Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12
- PMID: 9705952
- PMCID: PMC2213347
- DOI: 10.1084/jem.188.4.699
Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12
Abstract
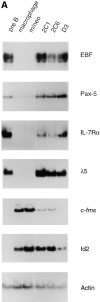
The transcription factors encoded by the E2A and early B cell factor (EBF) genes are required for the proper development of B lymphocytes. However, the absence of B lineage cells in E2A- and EBF-deficient mice has made it difficult to determine the function or relationship between these proteins. We report the identification of a novel model system in which the role of E2A and EBF in the regulation of multiple B lineage traits can be studied. We found that the conversion of 70Z/3 pre-B lymphocytes to cells with a macrophage-like phenotype is associated with the loss of E2A and EBF. Moreover, we show that ectopic expression of the E2A protein E12 in this macrophage line results in the induction of many B lineage genes, including EBF, IL7Ralpha, lambda5, and Rag-1, and the ability to induce kappa light chain in response to mitogen. Activation of EBF may be one of the critical functions of E12 in regulating the B lineage phenotype since expression of EBF alone leads to the activation of a subset of E12-inducible traits. Our data demonstrate that, in the context of this macrophage line, E12 induces expression of EBF and together these transcription factors coordinately regulate numerous B lineage-associated genes.
Figures



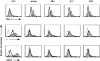
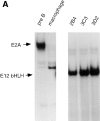






References
-
- Till JE, McCulloch EA. Hemopoietic stem cell differentiation. Biochim Biophys Acta. 1980;605:431–459. - PubMed
-
- Billips LG, Petitte D, Dorshkind K, Narayanan R, Chiu C-P, Landreth KS. Differential roles of stromal cells, interleukin-7, and kit-ligand in the regulation of B lymphopoiesis. Blood. 1992;79:1185–1192. - PubMed
-
- Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. - PubMed
-
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

