Identification of an UP element consensus sequence for bacterial promoters
- PMID: 9707549
- PMCID: PMC21410
- DOI: 10.1073/pnas.95.17.9761
Identification of an UP element consensus sequence for bacterial promoters
Abstract
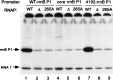
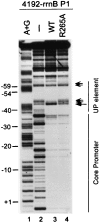
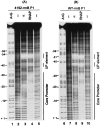
The UP element, a component of bacterial promoters located upstream of the -35 hexamer, increases transcription by interacting with the RNA polymerase alpha-subunit. By using a modification of the SELEX procedure for identification of protein-binding sites, we selected in vitro and subsequently screened in vivo for sequences that greatly increased promoter activity when situated upstream of the Escherichia coli rrnB P1 core promoter. A set of 31 of these upstream sequences increased transcription from 136- to 326-fold in vivo, considerably more than the natural rrnB P1 UP element, and was used to derive a consensus sequence: -59 nnAAA(A/T)(A/T)T(A/T)TTTTnnAAAAnnn -38. The most active selected sequence contained the derived consensus, displayed all of the properties of an UP element, and the interaction of this sequence with the alpha C-terminal domain was similar to that of previously characterized UP elements. The identification of the UP element consensus should facilitate a detailed understanding of the alpha-DNA interaction. Based on the evolutionary conservation of the residues in alpha responsible for interaction with UP elements, we suggest that the UP element consensus sequence should be applicable throughout eubacteria, should generally facilitate promoter prediction, and may be of use for biotechnological applications.
Figures


References
-
- Dombroski A J, Walter W A, Record M T J, Siegele D A, Gross C A. Cell. 1992;70:501–512. - PubMed
-
- Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. Science. 1993;262:1407–1413. - PubMed
-
- Blatter E E, Ross W, Tang H, Gourse R L, Ebright R H. Cell. 1994;78:889–896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

