Determination of transmembrane topology of an inward-rectifying potassium channel from Arabidopsis thaliana based on functional expression in Escherichia coli
- PMID: 9707551
- PMCID: PMC21412
- DOI: 10.1073/pnas.95.17.9773
Determination of transmembrane topology of an inward-rectifying potassium channel from Arabidopsis thaliana based on functional expression in Escherichia coli
Abstract
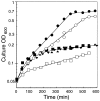
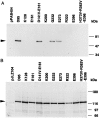
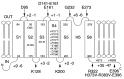
We report here that the inward-rectifying potassium channels KAT1 and AKT2 were functionally expressed in K+ uptake-deficient Escherichia coli. Immunological assays showed that KAT1 was translocated into the cell membrane of E. coli. Functional assays suggested that KAT1 was inserted topologically correctly into the cell membrane. In control experiments, the inactive point mutation in KAT1, T256R, did not complement for K+ uptake in E. coli. The inward-rectifying K+ channels of plants share a common hydrophobic domain comprising at least six membrane-spanning segments (S1-S6). The finding that a K+ channel can be expressed in bacteria was further exploited to determine the KAT1 membrane topology by a gene fusion approach using the bacterial reporter enzymes, alkaline phosphatase, which is active only in the periplasm, and beta-galactosidase. The enzyme activity from the alkaline phosphatase and beta-galactosidase fusion plasmid showed that the widely predicted S1, S2, S5, and S6 segments were inserted into the membrane. Although the S3 segment in the alkaline phosphatase fusion protein could not function as an export signal, the replacement of a negatively charged residue inside S3 with a neutral amino acid resulted in an increase in alkaline phosphatase activity, which indicates that the alkaline phosphatase was translocated into the periplasm. For membrane translocation of S3, the neutralization of a negatively charged residue in S3 may be required presumably because of pairing with a positively charged residue of S4. These results revealed that KAT1 has the common six transmembrane-spanning membrane topology that has been predicted for the Shaker superfamily of voltage-dependent K+ channels. Furthermore, the functional complementation of a bacterial K+ uptake mutant in this study is shown to be an alternative expression system for plant K+ channel proteins and a potent tool for their topological analysis.
Figures

Similar articles
-
Amino terminus and the first four membrane-spanning segments of the Arabidopsis K+ channel KAT1 confer inward-rectification property of plant-animal chimeric channels.J Biol Chem. 1995 Jul 28;270(30):17697-701. J Biol Chem. 1995. PMID: 7629068
-
Molecular dissection of the contribution of negatively and positively charged residues in S2, S3, and S4 to the final membrane topology of the voltage sensor in the K+ channel, KAT1.J Biol Chem. 2003 Apr 11;278(15):13227-34. doi: 10.1074/jbc.M300431200. Epub 2003 Jan 29. J Biol Chem. 2003. PMID: 12556517
-
Integration of Shaker-type K+ channel, KAT1, into the endoplasmic reticulum membrane: synergistic insertion of voltage-sensing segments, S3-S4, and independent insertion of pore-forming segments, S5-P-S6.Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):60-5. doi: 10.1073/pnas.012399799. Epub 2001 Dec 26. Proc Natl Acad Sci U S A. 2002. PMID: 11756658 Free PMC article.
-
Structure and function of potassium channels in plants: some inferences about the molecular origin of inward rectification in KAT1 channels (Review).Mol Membr Biol. 2003 Jan-Mar;20(1):19-25. doi: 10.1080/0968768021000057371. Mol Membr Biol. 2003. PMID: 12745922 Review.
-
Escherichia coli as an expression system for K(+) transport systems from plants.Am J Physiol Cell Physiol. 2001 Sep;281(3):C733-9. doi: 10.1152/ajpcell.2001.281.3.C733. Am J Physiol Cell Physiol. 2001. PMID: 11502550 Review.
Cited by
-
Heterelogous expression of plant genes.Int J Plant Genomics. 2009;2009:296482. doi: 10.1155/2009/296482. Epub 2009 Aug 6. Int J Plant Genomics. 2009. PMID: 19672459 Free PMC article.
-
Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis.Plant Mol Biol. 2000 Jul;43(4):515-25. doi: 10.1023/a:1006496402463. Plant Mol Biol. 2000. PMID: 11052203
-
Enhancement of Na(+) uptake currents, time-dependent inward-rectifying K(+) channel currents, and K(+) channel transcripts by K(+) starvation in wheat root cells.Plant Physiol. 2000 Apr;122(4):1387-97. doi: 10.1104/pp.122.4.1387. Plant Physiol. 2000. PMID: 10759535 Free PMC article.
-
AtKC1, a silent Arabidopsis potassium channel alpha -subunit modulates root hair K+ influx.Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):4079-84. doi: 10.1073/pnas.052677799. Proc Natl Acad Sci U S A. 2002. PMID: 11904452 Free PMC article.
-
Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa.Plant Mol Biol. 2002 Nov;50(4-5):623-33. doi: 10.1023/a:1019951023362. Plant Mol Biol. 2002. PMID: 12374296
References
-
- Schroeder J I, Ward J M, Gassmann W. Annu Rev Biophys Biomol Struct. 1994;23:441–471. - PubMed
-
- Maathius F J M, Sanders D. Physiol Plant. 1996;96:158–168.
-
- Kochian L V, Lucas W J. Adv Not Res. 1988;15:93–178.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

