New enzyme lineages by subdomain shuffling
- PMID: 9707558
- PMCID: PMC21419
- DOI: 10.1073/pnas.95.17.9813
New enzyme lineages by subdomain shuffling
Abstract
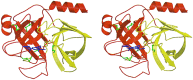
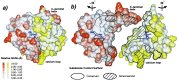
Protein functions have evolved in part via domain recombination events. Such events, for example, recombine structurally independent functional domains and shuffle targeting, regulatory, and/or catalytic functions. Domain recombination, however, can generate new functions, as implied by the observation of catalytic sites at interfaces of distinct folding domains. If useful to an evolving organism, such initially rudimentary functions would likely acquire greater efficiency and diversity, whereas the initially distinct folding domains would likely develop into single functional domains. This represents the probable evolution of the S1 serine protease family, whose two homologous beta-barrel subdomains assemble to form the binding sites and the catalytic machinery. Among S1 family members, the contact interface and catalytic residues are highly conserved whereas surrounding surfaces are highly variable. This observation suggests a new strategy to engineer viable proteins with novel properties, by swapping folding subdomains chosen from among protein family members. Such hybrid proteins would retain properties conserved throughout the family, including folding stability as single domain proteins, while providing new surfaces amenable to directed evolution or engineering of specific new properties. We show here that recombining the N-terminal subdomain from coagulation factor X with the C-terminal subdomain from trypsin creates a potent enzyme (fXYa) with novel properties, in particular a broad substrate specificity. As shown by the 2.15-A crystal structure, plasticity at the hydrophobic subdomain interface maintains activity, while surface loops are displaced compared with the parent subdomains. fXYa thus represents a new serine proteinase lineage with hybrid fX, trypsin, and novel properties.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

