Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant
- PMID: 9707568
- PMCID: PMC21429
- DOI: 10.1073/pnas.95.17.9873
Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant
Abstract
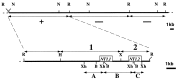
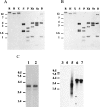
All parasitic protozoa studied to date are incapable of purine biosynthesis and must therefore salvage purine nucleobases or nucleosides from their hosts. This salvage process is initiated by purine transporters on the parasite cell surface. We have used a mutant line (TUBA5) of Leishmania donovani that is deficient in adenosine/pyrimidine nucleoside transport activity (LdNT1) to clone genes encoding these nucleoside transporters by functional rescue. Two such genes, LdNT1.1 and LdNT1.2, have been sequenced and shown to encode deduced polypeptides with significant sequence identity to the human facilitative nucleoside transporter hENT1. Hydrophobicity analysis of the LdNT1.1 and LdNT1.2 proteins predicted 11 transmembrane domains. Transfection of the adenosine/pyrimidine nucleoside transport-deficient TUBA5 parasites with vectors containing the LdNT1.1 and LdNT1.2 genes confers sensitivity to the cytotoxic adenosine analog tubercidin and concurrently restores the ability of this mutant line to take up [3H]adenosine and [3H]uridine. Moreover, expression of the LdNT1.2 ORF in Xenopus oocytes significantly increases their ability to take up [3H]adenosine, confirming that this single protein is sufficient to mediate nucleoside transport. These results establish genetically and biochemically that both LdNT1 genes encode functional adenosine/pyrimidine nucleoside transporters.
Figures



Similar articles
-
Point mutations in a nucleoside transporter gene from Leishmania donovani confer drug resistance and alter substrate selectivity.Proc Natl Acad Sci U S A. 2001 May 22;98(11):6092-7. doi: 10.1073/pnas.101537298. Epub 2001 May 15. Proc Natl Acad Sci U S A. 2001. PMID: 11353834 Free PMC article.
-
Cloning of a novel inosine-guanosine transporter gene from Leishmania donovani by functional rescue of a transport-deficient mutant.J Biol Chem. 2000 Jul 7;275(27):20935-41. doi: 10.1074/jbc.M002418200. J Biol Chem. 2000. PMID: 10783393
-
Functional characterization of nucleoside transporter gene replacements in Leishmania donovani.Mol Biochem Parasitol. 2006 Dec;150(2):300-7. doi: 10.1016/j.molbiopara.2006.09.002. Epub 2006 Sep 27. Mol Biochem Parasitol. 2006. PMID: 17050001 Free PMC article.
-
Molecular genetics of nucleoside transporters in Leishmania and African trypanosomes.Biochem Pharmacol. 2001 Jul 15;62(2):149-55. doi: 10.1016/s0006-2952(01)00663-3. Biochem Pharmacol. 2001. PMID: 11389872 Review.
-
Nucleoside transporters of parasitic protozoa.Trends Parasitol. 2001 Mar;17(3):142-5. doi: 10.1016/s1471-4922(00)01806-7. Trends Parasitol. 2001. PMID: 11286799 Review.
Cited by
-
An ab Initio structural model of a nucleoside permease predicts functionally important residues.J Biol Chem. 2009 Jul 10;284(28):19067-76. doi: 10.1074/jbc.M109.017947. Epub 2009 May 8. J Biol Chem. 2009. PMID: 19429678 Free PMC article.
-
The Trypanosoma cruzi TcrNT2 Nucleoside Transporter Is a Conduit for the Uptake of 5-F-2'-Deoxyuridine and Tubercidin Analogues.Molecules. 2022 Nov 19;27(22):0. doi: 10.3390/molecules27228045. Molecules. 2022. PMID: 36432150 Free PMC article.
-
Characterization of a Novel Endoplasmic Reticulum Protein Involved in Tubercidin Resistance in Leishmania major.PLoS Negl Trop Dis. 2016 Sep 8;10(9):e0004972. doi: 10.1371/journal.pntd.0004972. eCollection 2016 Sep. PLoS Negl Trop Dis. 2016. PMID: 27606425 Free PMC article.
-
Role of transmembrane domain 4 in ligand permeation by Crithidia fasciculata equilibrative nucleoside transporter 2 (CfNT2).J Biol Chem. 2010 Feb 26;285(9):6024-35. doi: 10.1074/jbc.M109.074351. Epub 2009 Dec 26. J Biol Chem. 2010. PMID: 20037157 Free PMC article.
-
Nucleoside and nucleobase transporters in parasitic protozoa.Eukaryot Cell. 2004 Apr;3(2):245-54. doi: 10.1128/EC.3.2.245-254.2004. Eukaryot Cell. 2004. PMID: 15075255 Free PMC article. Review. No abstract available.
References
-
- Walsh J A, Warren K S. N Engl J Med. 1979;301:967–974. - PubMed
-
- Lainson R, Shaw J J. Nature (London) 1978;273:595–600. - PubMed
-
- Berens R L, Krug E C, Marr J J. In: Purine and Pyrimidine Metabolism. Marr J J, Müller M, editors. New York: Academic; 1995. pp. 89–117.
-
- Marr J J, Berens R L. Mol Biochem Parasitol. 1983;7:339–356. - PubMed
-
- Aronow B, Kaur K, McCartan K, Ullman B. Mol Biochem Parasitol. 1987;22:29–37. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

