A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots
- PMID: 9707576
- PMCID: PMC21437
- DOI: 10.1073/pnas.95.17.9920
A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots
Abstract
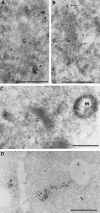
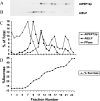
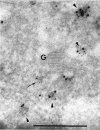
Targeting of protein cargo to the vacuole/lysosome is a multistep process that appears to have conserved features between mammalian, yeast, and plant cells. In each case, some soluble vacuolar/lysosomal proteins are believed to be bound by transmembrane cargo receptors in the trans-Golgi network (TGN) that redirect these proteins into clathrin-coated vesicles. These vesicles then appear to be transported to the prevacuole/endosome by a trafficking machinery that requires components identified in other vesicle-targeting steps such as N-ethylmaleimide-sensitive factor (NSF), soluble NSF attachment protein (SNAP), SNAP receptors (SNAREs), rab-type GTPases, and Sec1p homologs. Two likely members of this trafficking machinery have been characterized from Arabidopsis thaliana: AtPEP12p, a t-SNARE that resides on a what we now call a prevacuolar compartment, and AtELP, a protein that shares many common features with mammalian and yeast transmembrane cargo receptors. Here, we have further investigated the intracellular distribution of AtELP. We have found that AtELP is located at the trans-Golgi of Arabidopsis root cells, and that its C terminus can preferentially interact in vitro with the mammalian TGN-specific AP-1 clathrin-adapter complex, suggesting a likely role in clathrin-coated, vesicle-directed trafficking at the TGN. Further, consistent with a role in trafficking of vacuolar cargo, we have found that AtELP partially colocalizes with AtPEP12p on a prevacuolar compartment.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

