Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis
- PMID: 9707643
- PMCID: PMC21504
- DOI: 10.1073/pnas.95.17.10306
Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis
Abstract
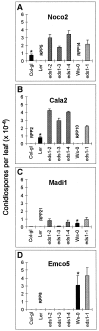
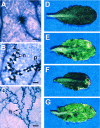
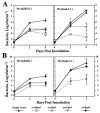
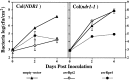
The Arabidopsis genes EDS1 and NDR1 were shown previously by mutational analysis to encode essential components of race-specific disease resistance. Here, we examined the relative requirements for EDS1 and NDR1 by a broad spectrum of Resistance (R) genes present in three Arabidopsis accessions (Columbia, Landsberg-erecta, and Wassilewskija). We show that there is a strong requirement for EDS1 by a subset of R loci (RPP2, RPP4, RPP5, RPP21, and RPS4), conferring resistance to the biotrophic oomycete Peronospora parasitica, and to Pseudomonas bacteria expressing the avirulence gene avrRps4. The requirement for NDR1 by these EDS1-dependent R loci is either weak or not measurable. Conversely, three NDR1-dependent R loci, RPS2, RPM1, and RPS5, operate independently of EDS1. Another RPP locus, RPP8, exhibits no strong exclusive requirement for EDS1 or NDR1 in isolate-specific resistance to P. parasitica, although resistance is compromised weakly by eds1. Similarly, resistance conditioned by two EDS1-dependent RPP genes, RPP4 and RPP5, is impaired partially by ndr1, implicating a degree of pathway cross-talk. Our results provide compelling evidence for the preferential utilization of either signaling component by particular R genes and thus define at least two disease resistance pathways. The data also suggest that strong dependence on EDS1 or NDR1 is governed by R protein structural type rather than pathogen class.
Figures
References
-
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. - PubMed
-
- Zhou J, Loh Y-T, Bressan R A, Martin G B. Cell. 1995;83:925–935. - PubMed
-
- Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. - PubMed
-
- Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. - PubMed
-
- Jones D A, Jones J D G. Adv Bot Res Inc Adv Plant Pathol. 1996;24:89–167.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

