CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif
- PMID: 9710584
- PMCID: PMC109085
- DOI: 10.1128/MCB.18.9.5000
CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif
Abstract
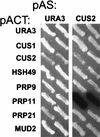
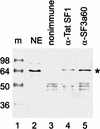
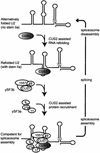
A screen for suppressors of a U2 snRNA mutation identified CUS2, an atypical member of the RNA recognition motif (RRM) family of RNA binding proteins. CUS2 protein is associated with U2 RNA in splicing extracts and interacts with PRP11, a subunit of the conserved splicing factor SF3a. Absence of CUS2 renders certain U2 RNA folding mutants lethal, arguing that a normal activity of CUS2 is to help refold U2 into a structure favorable for its binding to SF3b and SF3a prior to spliceosome assembly. Both CUS2 function in vivo and the in vitro RNA binding activity of CUS2 are disrupted by mutation of the first RRM, suggesting that rescue of misfolded U2 involves the direct binding of CUS2. Human Tat-SF1, reported to stimulate Tat-specific, transactivating region-dependent human immunodeficiency virus transcription in vitro, is structurally similar to CUS2. Anti-Tat-SF1 antibodies coimmunoprecipitate SF3a66 (SAP62), the human homolog of PRP11, suggesting that Tat-SF1 has a parallel function in splicing in human cells.
Figures





References
-
- Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. - PubMed
-
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ares M, Jr, Igel A H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
