The ability of CD40L, but not lipopolysaccharide, to initiate immunoglobulin switching to immunoglobulin G1 is explained by differential induction of NF-kappaB/Rel proteins
- PMID: 9710636
- PMCID: PMC109137
- DOI: 10.1128/MCB.18.9.5523
The ability of CD40L, but not lipopolysaccharide, to initiate immunoglobulin switching to immunoglobulin G1 is explained by differential induction of NF-kappaB/Rel proteins
Abstract
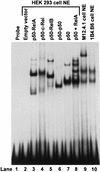
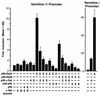
Antibodies of the immunoglobulin G1 class are induced in mice by T-cell-dependent antigens but not by lipopolysaccharide (LPS). CD40 engagement contributes to this preferential isotype production by activating NF-kappaB/Rel to induce germ line gamma1 transcripts, which are essential for class switch recombination. Although LPS also activates NF-kappaB, it poorly induces germ line gamma1 transcripts. Western blot analyses show that CD40 ligand (CD40L) induces all NF-kappaB/Rel proteins, whereas LPS activates predominantly p50 and c-Rel. Electrophoretic mobility shift assays show that in CD40L-treated cells, p50-RelA and p50-RelB dimers are the major NF-kappaB complexes binding to the germ line gamma1 promoter, whereas in LPS-treated cells, p50-c-Rel and p50-p50 dimers are the major binding complexes. Transfection of expression plasmids for NF-kappaB/Rel fusion proteins (forced dimers) indicates that p50-RelA and p50-RelB dimers activate the germ line gamma1 promoter and that p50-c-Rel and p50-p50 dimers inhibit this activation by competitively binding to the promoter without activating the promoter. Therefore, germ line gamma1 transcription depends on the composition of NF-kappaB/Rel proteins.
Figures






References
-
- Baldwin A S., Jr The NF-κB and I-κB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
-
- Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. - PubMed
-
- Berton M T, Linehan L A. IL-4 activates a latent DNA-binding factor that binds a shared IFN-γ and IL-4 response element present in the germ-line γ1 Ig promoter. J Immunol. 1995;154:4513–4525. - PubMed
-
- Brasier A, Tate J, Habener J. Optimized use of the firefly luciferase assay as a reporter gene in mammalian cell lines. BioTechniques. 1989;7:1116–1122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
