Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5
- PMID: 9712774
- PMCID: PMC108512
- DOI: 10.1128/IAI.66.9.4244-4253.1998
Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5
Abstract
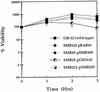
In order to determine the importance of the O75 O antigen versus the K5 capsular antigen and the bimodal distribution of lipopolysaccharides (LPSs) in protection from complement-mediated lysis, mutants were made by insertion of a cat or an aphA gene in or in place of genes necessary for the synthesis of LPS and/or the K antigen of an O75(+) K5(+) uropathogenic Escherichia coli strain, GR-12. Mutations were made in the following genes: the rfbD gene (required for the synthesis of TDP-rhamnose), the rfbKM genes (necessary for the synthesis of GDP-mannose), the rol gene (regulating O-antigen length), the kfiC gene (encoding a putative glycosyltransferase), and the kfiC-rfbD genes. The resulting phenotypes were rough (O75(-)), core plus one partial O-antigen subunit, random distribution of O-antigen chain lengths, acapsular (K5(-)), and O75(-) K5(-), respectively. All five mutants and GR-12 were analyzed for survival in 80% serum. The GR-12 parent was resistant, exhibiting a 500% increase in numbers. The rol, rfbKM, rfbD, and kfiC-rfbD mutants were sensitive, experiencing 99%, 99.9%, 99.9%, and at least 99.999% killing, respectively, in the first hour. The kfiC mutant, however, increased in numbers in the first hour but experienced delayed sensitivity, decreasing in viability by 80% in the third hour. Single mutants were complemented with the wild-type gene in trans, showing restoration of the wild-type phenotype and serum resistance. Therefore, the O75 antigen is more important for survival in serum than the K5 antigen, and regulation of the O75 O-antigen chain length is crucial for protection of the bacteria from complement-mediated lysis.
Figures






References
-
- Batchelor R A. Ph.D. thesis. Houston, Tex: Baylor College of Medicine; 1992.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

