Parallel and divergent genotypic evolution in experimental populations of Ralstonia sp
- PMID: 9721265
- PMCID: PMC107437
- DOI: 10.1128/JB.180.17.4325-4331.1998
Parallel and divergent genotypic evolution in experimental populations of Ralstonia sp
Abstract
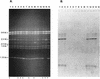
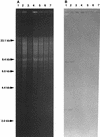
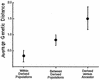
Genetic rearrangements within a population of bacteria were analyzed to understand the degree of divergence occurring after experimental evolution. We used 18 replicate populations founded from Ralstonia sp. strain TFD41 that had been propagated for 1,000 generations with 2,4-dichlorophenoxyacetic acid (2,4-D) as the carbon source. Genetic divergence was examined by restriction fragment length polymorphism analysis of the incumbent plasmid that carries the 2,4-D catabolic genes and by amplification of random regions of the genome via PCR. In 18 evolved clones examined, we observed duplication within the plasmid, including the tfdA gene, which encodes a 2,4-D dioxygenase that catalyzes the first step in the 2,4-D catabolic pathway. In 71 of 72 evolved clones, a common 2.4-kb PCR product was lost when genomic fingerprints produced by PCR amplification using degenerate primers based on repetitive extragenic palindromic (REP) sequences (REP-PCR) were compared. The nucleotide sequence of the 2.4-kb PCR product has homology to the TRAP (tripartite ATP-independent periplasmic) solute transporter gene family. Hybridization of the 2. 4-kb REP-PCR product from the ancestor to genomic DNA from the evolved populations showed that the loss of the PCR product resulted from deletions in the genome. Deletions in the plasmid and presence and/or absence of other REP-PCR products were also found in these clones but at much lower frequencies. The common and uncommon genetic changes observed show that both parallel and divergent genotypic evolution occurred in replicate populations of this bacterium.
Figures


References
-
- Ajioka J W, Hartl D L. Population dynamics of transposable elements. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: ASM Press; 1989. pp. 939–958.
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arber W. Elements in microbial evolution. J Mol Evol. 1991;33:4–12. - PubMed
-
- Arber W. Evolution of prokaryotic genomes. Gene. 1993;135:49–56. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

