Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100
- PMID: 9721268
- PMCID: PMC107440
- DOI: 10.1128/JB.180.17.4344-4349.1998
Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100
Abstract
Cytosolic extracts of Lactobacillus johnsonii 100-100 (previously reported as Lactobacillus sp. strain 100-100) contain four heterotrimeric isozymes composed of two peptides, alpha and beta, with conjugated bile salt hydrolase (BSH) activity. We now report cloning, from the genome of strain 100-100, a 2,977-bp DNA segment that expresses BSH activity in Escherichia coli. The sequencing of this segment showed that it contained one complete and two partial open reading frames (ORFs). The 3' partial ORF (927 nucleotides) was predicted by BLAST and confirmed with 5' and 3' deletions to be a BSH gene. Thermal asymmetric interlaced PCR was used to extend and complete the 948-nucleotide sequence of the BSH gene 3' of the cloned segment. The predicted amino acid sequence of the 5' partial ORF (651 nucleotides) was about 80% similar to the C-terminal half of the largest, complete ORF (1,353 nucleotides), and these two putative proteins were similar to several amine, multidrug resistance, and sugar transport proteins of the major facilitator superfamily. E. coli DH5alpha cells transformed with a construct containing these ORFs, in concert with an extracellular factor produced by strain 100-100, demonstrated levels of uptake of [14C]taurocholic acid that were increased as much as threefold over control levels. [14C]Cholic acid was taken up in similar amounts by strain DH5alpha pSportI (control) and DH5alpha p2000 (transport clones). These findings support a hypothesis that the ORFs are conjugated bile salt transport genes which may be arranged in an operon with BSH genes.
Figures
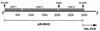




References
-
- Baron S F, Hylemon P B. Biotransformation of bile acids, cholesterol, and steroid hormones. In: Mackie R I, White B A, editors. Gastrointestinal microbiology. I. Gastrointestinal ecosystems and fermentations. New York, N.Y: International Thomson Publishing; 1997. pp. 470–510.
-
- Batta A K, Salen G, Arora R, Shefer S, Batta M, Person A. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J Biol Chem. 1990;265:10925–10928. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

