Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. Strain HR199 and molecular characterization of the gene
- PMID: 9721273
- PMCID: PMC107445
- DOI: 10.1128/JB.180.17.4387-4391.1998
Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. Strain HR199 and molecular characterization of the gene
Abstract
The coniferyl aldehyde dehydrogenase (CALDH) of Pseudomonas sp. strain HR199 (DSM7063), which catalyzes the NAD+-dependent oxidation of coniferyl aldehyde to ferulic acid and which is induced during growth with eugenol as the carbon source, was purified and characterized. The native protein exhibited an apparent molecular mass of 86,000 +/- 5,000 Da, and the subunit mass was 49.5 +/- 2.5 kDa, indicating an alpha2 structure of the native enzyme. The optimal oxidation of coniferyl aldehyde to ferulic acid was obtained at a pH of 8.8 and a temperature of 26 degreesC. The Km values for coniferyl aldehyde and NAD+ were about 7 to 12 microM and 334 microM, respectively. The enzyme also accepted other aromatic aldehydes as substrates, whereas aliphatic aldehydes were not accepted. The NH2-terminal amino acid sequence of CALDH was determined in order to clone the encoding gene (calB). The corresponding nucleotide sequence was localized on a 9.4-kbp EcoRI fragment (E94), which was subcloned from a Pseudomonas sp. strain HR199 genomic library in the cosmid pVK100. The partial sequencing of this fragment revealed an open reading frame of 1,446 bp encoding a protein with a relative molecular weight of 51,822. The deduced amino acid sequence, which is reported for the first time for a structural gene of a CALDH, exhibited up to 38.5% amino acid identity (60% similarity) to NAD+-dependent aldehyde dehydrogenases from different sources.
Figures
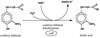


References
-
- Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. - PubMed
-
- Boyed L A, Adam L, Pelcher L E, McHughen A, Hirji R, Selvaraj G. Characterization of an Escherichia coli gene encoding betaine aldehyde dehydrogenase (BADH): structural similarity to mammalian ALDHs and plant BADH. Gene. 1991;103:45–52. - PubMed
-
- Bullock W O, Fernandez J M, Stuart J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

