Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate
- PMID: 9721279
- PMCID: PMC107451
- DOI: 10.1128/JB.180.17.4426-4434.1998
Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate
Abstract
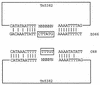
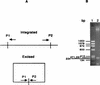
Mechanisms for the intercellular transfer of VanB-type vancomycin resistance determinants and for the almost universal association of these determinants with those for high-level ampicillin resistance remain poorly defined. We report the discovery of Tn5382, a ca. 27-kb putative transposon encoding VanB-type glycopeptide resistance in Enterococcus faecium. Open reading frames internal to the right end of Tn5382 and downstream of the vanXB dipeptidase gene exhibit significant homology to genes encoding the excisase and integrase of conjugative transposon Tn916. The ends of Tn5382 are also homologous to the ends of Tn916, especially in regions bound by the integrase enzyme. PCR amplification experiments indicate that Tn5382 excises to form a circular intermediate in E. faecium. Integration of Tn5382 in the chromosome of E. faecium C68 has occurred 113 bp downstream of the stop codon for the pbp5 gene, which encodes high-level ampicillin resistance in this clinical isolate. Transfer of vancomycin, ampicillin, and tetracycline resistance from C68 to an E. faecium recipient strain occurs at low frequency in vitro and is associated with acquisition of a 130- to 160-kb segment of DNA that contains Tn5382, the pbp5 gene, and its putative repressor gene, psr. The interenterococcal transfer of this large chromosomal element appears to be the primary mechanism for vanB operon spread in northeast Ohio. These results expand the known family of Tn916-related transposons, suggest a mechanism for vanB operon entry into and dissemination among enterococci, and provide an explanation for the nearly universal association of vancomycin and high-level ampicillin resistance in clinical E. faecium strains.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

