SdeK is required for early fruiting body development in Myxococcus xanthus
- PMID: 9721305
- PMCID: PMC107477
- DOI: 10.1128/JB.180.17.4628-4637.1998
SdeK is required for early fruiting body development in Myxococcus xanthus
Abstract
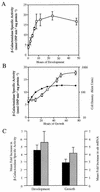
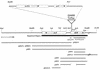
Myxococcus xanthus cells carrying the Omega4408 Tn5lac insertion at the sde locus show defects in fruiting body development and sporulation. Our analysis of sde expression patterns showed that this locus is induced early in the developmental program (0 to 2 h) and that expression increases approximately fivefold after 12 h of development. Further studies showed that expression of sde is induced as growing cells enter stationary phase, suggesting that activation of the sde locus is not limited to the developmental process. Because the peak levels of sde expression in both an sde+ and an sde mutant background were similar, we conclude that the sde locus is not autoregulated. Characterization of the sde locus by DNA sequence analysis indicated that the Omega4408 insertion occurred within the sdeK gene. Primer extension analyses localized the 5' end of sde transcript to a guanine nucleotide 307 bp upstream of the proposed start for the SdeK coding sequence. The DNA sequence in the -12 and -24 regions upstream of the sde transcriptional start site shows similarity to the sigma54 family of promoters. The results of complementation studies suggest that the defects in development and sporulation caused by the Omega4408 insertion are due to an inactivation of sdeK. The predicted amino acid sequence of SdeK was found to have similarity to the sequences of the histidine protein kinases of two-component regulatory systems. Based on our results, we propose that SdeK may be part of a signal transduction pathway required for the activation and propagation of the early developmental program.
Figures




References
-
- Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. - PubMed
-
- Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein coding sequences. Gene. 1984;30:157–166. - PubMed
-
- Blanco G, Drummond M, Woodley P, Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993;9:869–879. - PubMed
-
- David M, Daveran M L, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

