Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE
- PMID: 9722605
- PMCID: PMC2132875
- DOI: 10.1083/jcb.142.4.913
Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE
Abstract
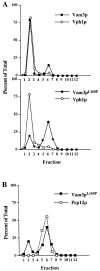
The transport of newly synthesized proteins through the vacuolar protein sorting pathway in the budding yeast Saccharomyces cerevisiae requires two distinct target SNAP receptor (t-SNARE) proteins, Pep12p and Vam3p. Pep12p is localized to the pre-vacuolar endosome and its activity is required for transport of proteins from the Golgi to the vacuole through a well defined route, the carboxypeptidase Y (CPY) pathway. Vam3p is localized to the vacuole where it mediates delivery of cargoes from both the CPY and the recently described alkaline phosphatase (ALP) pathways. Surprisingly, despite their organelle-specific functions in sorting of vacuolar proteins, overexpression of VAM3 can suppress the protein sorting defects of pep12Delta cells. Based on this observation, we developed a genetic screen to identify domains in Vam3p (e.g., localization and/or specific protein-protein interaction domains) that allow it to efficiently substitute for Pep12p. Using this screen, we identified mutations in a 7-amino acid sequence in Vam3p that lead to missorting of Vam3p from the ALP pathway into the CPY pathway where it can substitute for Pep12p at the pre-vacuolar endosome. This region contains an acidic di-leucine sequence that is closely related to sorting signals required for AP-3 adaptor-dependent transport in both yeast and mammalian systems. Furthermore, disruption of AP-3 function also results in the ability of wild-type Vam3p to compensate for pep12 mutants, suggesting that AP-3 mediates the sorting of Vam3p via the di-leucine signal. Together, these data provide the first identification of an adaptor protein-specific sorting signal in a t-SNARE protein, and suggest that AP-3-dependent sorting of Vam3p acts to restrict its interaction with compartment-specific accessory proteins, thereby regulating its function. Regulated transport of cargoes such as Vam3p through the AP-3-dependent pathway may play an important role in maintaining the unique composition, function, and morphology of the vacuole.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

