Quantitative assessment of enzyme specificity in vivo: P2 recognition by Kex2 protease defined in a genetic system
- PMID: 9724712
- PMCID: PMC27903
- DOI: 10.1073/pnas.95.18.10384
Quantitative assessment of enzyme specificity in vivo: P2 recognition by Kex2 protease defined in a genetic system
Abstract
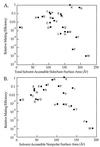
The specificity of the yeast proprotein-processing Kex2 protease was examined in vivo by using a sensitive, quantitative assay. A truncated prepro-alpha-factor gene encoding an alpha-factor precursor with a single alpha-factor repeat was constructed with restriction sites for cassette mutagenesis flanking the single Kex2 cleavage site (-SLDKR downward arrowEAEA-). All of the 19 substitutions for the Lys (P2) residue in the cleavage site were made. The wild-type and mutant precursors were expressed in a yeast strain lacking the chromosomal genes encoding Kex2 and prepro-alpha-factor. Cleavage of the 20 sites by Kex2, expressed at the wild-type level, was assessed by using a quantitative-mating assay with an effective range greater than six orders of magnitude. All substitutions for Lys at P2 decreased mating, from 2-fold for Arg to >10(6)-fold for Trp. Eviction of the Kex2-encoding plasmid indicated that cleavage of mutant sites by other cellular proteases was not a complicating factor. Mating efficiencies of strains expressing the mutant precursors correlated well with the specificity (kcat/KM) of purified Kex2 for comparable model peptide substrates, validating the in vivo approach as a quantitative method. The results support the conclusion that KM, which is heavily influenced by the nature of the P2 residue, is a major determinant of cleavage efficiency in vivo. P2 preference followed the rank order: Lys > Arg > Thr > Pro > Glu > Ile > Ser > Ala > Asn > Val > Cys > AsP > Gln > Gly > His > Met > Leu > Tyr > Phe > Trp.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

