Tn5/IS50 target recognition
- PMID: 9724770
- PMCID: PMC27961
- DOI: 10.1073/pnas.95.18.10716
Tn5/IS50 target recognition
Abstract
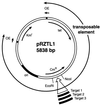
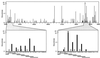
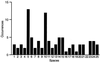
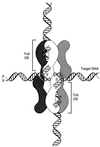
This communication reports an analysis of Tn5/IS50 target site selection by using an extensive collection of Tn5 and IS50 insertions in two relatively small regions of DNA (less than 1 kb each). For both regions data were collected resulting from in vitro and in vivo transposition events. Since the data sets are consistent and transposase was the only protein present in vitro, this demonstrates that target selection is a property of only transposase. There appear to be two factors governing target selection. A target consensus sequence, which presumably reflects the target selection of individual pairs of Tn5/IS50 bound transposase protomers, was deduced by analyzing all insertion sites. The consensus Tn5/IS50 target site is A-GNTYWRANC-T. However, we observed that independent insertion sites tend to form groups of closely located insertions (clusters), and insertions very often were spaced in a 5-bp periodic fashion. This suggests that Tn5/IS50 target selection is facilitated by more than two transposase protomers binding to the DNA, and, thus, for a site to be a good target, the overlapping neighboring DNA should be a good target, too. Synthetic target sequences were designed and used to test and confirm this model.
Figures

References
-
- Galas D J, Chandler M. In: Mobile DNA. Berg D E, Howe M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 109–162.
-
- Berg D E. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 109–162.
-
- Bukhari A I, Zipser D. Nature (London) 1972;236:240–243.
-
- Lichtenstein C, Brenner S. Nature (London) 1982;297:601–603. - PubMed
-
- Klaer R, Kühn S, Tillmann E, Fritz H-J, Starlinger P. Mol Gen Genet. 1981;181:169–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

