Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells
- PMID: 9724779
- PMCID: PMC27970
- DOI: 10.1073/pnas.95.18.10769
Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells
Abstract
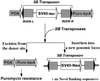
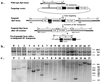
Mouse has become an increasingly important organism for modeling human diseases and for determining gene function in a mammalian context. Unfortunately, transposon-tagged mutagenesis, one of the most valuable tools for functional genomics, still is not available in this organism. On the other hand, it has long been speculated that members of the Tc1/mariner-like elements may be less dependent on host factors and, hence, can be introduced into heterologous organisms. However, this prediction has not been realized in mice. We report here the chromosomal transposition of the Sleeping Beauty (SB) element in mouse embryonic stem cells, providing evidence that it can be used as an in vivo mutagen in mice.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

