ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex
- PMID: 9724795
- PMCID: PMC27986
- DOI: 10.1073/pnas.95.18.10860
ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex
Abstract
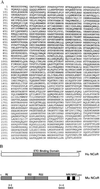
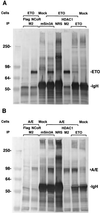
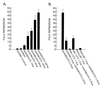
The t(8;21) translocation between two genes known as AML1 and ETO is seen in approximately 12-15% of all acute myeloid leukemia (AML) and is the second-most-frequently observed nonrandom genetic alteration associated with AML. AML1 up-regulates a number of target genes critical to normal hematopoiesis, whereas the AML1/ETO fusion interferes with this trans-activation. We discovered that the fusion partner ETO binds to the human homolog of the murine nuclear receptor corepressor (N-CoR). The interaction is mediated by two unusual zinc finger motifs present at the carboxyl terminus of ETO. Human N-CoR (HuN-CoR), which we cloned and sequenced in its entirety, encodes a 2,440-amino acid polypeptide and has a central domain that binds ETO. N-CoR, mammalian Sin3 (mSin3A and B), and histone deacetylase 1 (HDAC1) form a complex that alters chromatin structure and mediates transcriptional repression by nuclear receptors and by a number of oncoregulatory proteins. We found that ETO, through its interaction with the N-CoR/mSin3/HDAC1 complex, is also a potent repressor of transcription. This observation provides a mechanism for how the AML1/ETO fusion may inhibit expression of AML1-responsive target genes and disturb normal hematopoiesis.
Figures


References
-
- Look A T. Science. 1997;278:1059–1064. - PubMed
-
- Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Blood. 1992;80:1825–1831. - PubMed
-
- Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. Trends Genet. 1993;9:338–341. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

