Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis
- PMID: 9725903
- PMCID: PMC25507
- DOI: 10.1091/mbc.9.9.2407
Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis
Abstract
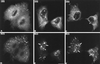
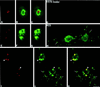
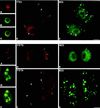
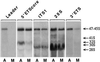
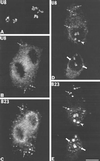
Previous studies showed that components implicated in pre-rRNA processing, including U3 small nucleolar (sno)RNA, fibrillarin, nucleolin, and proteins B23 and p52, accumulate in perichromosomal regions and in numerous mitotic cytoplasmic particles, termed nucleolus-derived foci (NDF) between early anaphase and late telophase. The latter structures were analyzed for the presence of pre-rRNA by fluorescence in situ hybridization using probes for segments of pre-rRNA with known half-lives. The NDF did not contain the short-lived 5'-external transcribed spacer (ETS) leader segment upstream from the primary processing site in 47S pre-rRNA. However, the NDF contained sequences from the 5'-ETS core, 18S, internal transcribed spacer 1 (ITS1), and 28S segments and also had detectable, but significantly reduced, levels of the 3'-ETS sequence. Northern analyses showed that in mitotic cells, the latter sequences were present predominantly in 45S-46S pre-rRNAs, indicating that high-molecular weight processing intermediates are preserved during mitosis. Two additional essential processing components were also found in the NDF: U8 snoRNA and hPop1 (a protein component of RNase MRP and RNase P). Thus, the NDF appear to be large complexes containing partially processed pre-rRNA associated with processing components in which processing has been significantly suppressed. The NDF may facilitate coordinated assembly of postmitotic nucleoli.
Figures



References
-
- Beven AF, Lee R, Razaz M, Leader DJ, Brown JWS, Shaw PJ. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J Cell Sci. 1996;109:1241–1251. - PubMed
-
- Bootsma D, Budke L, Vos O. Studies on synchronous division of tissue culture cells initiated by excess thymidine. Exp Cell Res. 1964;33:301–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

