Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola
- PMID: 9726857
- PMCID: PMC106707
- DOI: 10.1128/AEM.64.9.3180-3187.1998
Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola
Abstract
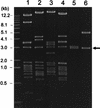
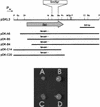
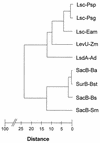
Plant-pathogenic bacteria produce various extracellular polysaccharides (EPSs) which may function as virulence factors in diseases caused by these bacteria. The EPS levan is synthesized by the extracellular enzyme levansucrase in Pseudomonas syringae, Erwinia amylovora, and other bacterial species. The lsc genes encoding levansucrase from P. syringae pv. glycinea PG4180 and P. syringae pv. phaseolicola NCPPB 1321 were cloned, and their nucleotide sequences were determined. Heterologous expression of the lsc gene in Escherichia coli was found in four and two genomic library clones of strains PG4180 and NCPPB 1321, respectively. A 3. 0-kb PstI fragment common to all six clones conferred levan synthesis on E. coli when further subcloned. Nucleotide sequence analysis revealed a 1,248-bp open reading frame (ORF) derived from PG4180 and a 1,296-bp ORF derived from NCPPB 1321, which were both designated lsc. Both ORFs showed high homology to the E. amylovora and Zymomonas mobilis lsc genes at the nucleic acid and deduced amino acid sequence levels. Levansucrase was not secreted into the supernatant but was located in the periplasmic fraction of E. coli harboring the lsc gene. Expression of lsc was found to be dependent on the vector-based Plac promoter, indicating that the native promoter of lsc was not functional in E. coli. Insertion of an antibiotic resistance cassette in the lsc gene abolished levan synthesis in E. coli. A PCR screening with primers derived from lsc of P. syringae pv. glycinea PG4180 allowed the detection of this gene in a number of related bacteria.
Figures




Similar articles
-
Genomic Distribution and Divergence of Levansucrase-Coding Genes in Pseudomonas syringae.Genes (Basel). 2012 Feb 10;3(1):115-37. doi: 10.3390/genes3010115. Genes (Basel). 2012. PMID: 24704846 Free PMC article.
-
Characterization and mutational analysis of three allelic lsc genes encoding levansucrase in Pseudomonas syringae.J Bacteriol. 2001 Jun;183(11):3282-92. doi: 10.1128/JB.183.11.3282-3292.2001. J Bacteriol. 2001. PMID: 11344135 Free PMC article.
-
Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in an hrp-dependent manner within tobacco cells.Mol Plant Microbe Interact. 1997 Jul;10(5):580-8. doi: 10.1094/MPMI.1997.10.5.580. Mol Plant Microbe Interact. 1997. PMID: 9204563
-
dnaK and the heat stress response of Pseudomonas syringae pv. glycinea.Mol Plant Microbe Interact. 1999 Jul;12(7):563-74. doi: 10.1094/MPMI.1999.12.7.563. Mol Plant Microbe Interact. 1999. PMID: 10478477
-
Pseudomonas savastanoi pv. savastanoi: some like it knot.Mol Plant Pathol. 2012 Dec;13(9):998-1009. doi: 10.1111/j.1364-3703.2012.00816.x. Epub 2012 Jul 17. Mol Plant Pathol. 2012. PMID: 22805238 Free PMC article. Review.
Cited by
-
Genomic Distribution and Divergence of Levansucrase-Coding Genes in Pseudomonas syringae.Genes (Basel). 2012 Feb 10;3(1):115-37. doi: 10.3390/genes3010115. Genes (Basel). 2012. PMID: 24704846 Free PMC article.
-
A type II protein secretory pathway required for levansucrase secretion by Gluconacetobacter diazotrophicus.J Bacteriol. 2004 Aug;186(15):5031-9. doi: 10.1128/JB.186.15.5031-5039.2004. J Bacteriol. 2004. PMID: 15262940 Free PMC article.
-
The conserved upstream region of lscB/C determines expression of different levansucrase genes in plant pathogen Pseudomonas syringae.BMC Microbiol. 2014 Mar 27;14:79. doi: 10.1186/1471-2180-14-79. BMC Microbiol. 2014. PMID: 24670199 Free PMC article.
-
Thermoregulated expression and characterization of an NAD(P)H-dependent 2-cyclohexen-1-one reductase in the plant pathogenic bacterium Pseudomonas syringae pv. glycinea.J Bacteriol. 1999 Feb;181(3):814-22. doi: 10.1128/JB.181.3.814-822.1999. J Bacteriol. 1999. PMID: 9922244 Free PMC article.
-
An improved, high-quality draft genome sequence of the Germination-Arrest Factor-producing Pseudomonas fluorescens WH6.BMC Genomics. 2010 Sep 28;11:522. doi: 10.1186/1471-2164-11-522. BMC Genomics. 2010. PMID: 20920191 Free PMC article.
References
-
- Arrieta J, Hernandez L, Coego A, Suarez V, Balmori E, Menendez C, Petit-Glatron M F, Chambert R, Selman-Housein G. Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiology. 1996;142:1077–1085. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

