Growth rate regulation of rRNA content of a marine synechococcus (Cyanobacterium) strain
- PMID: 9726881
- PMCID: PMC106731
- DOI: 10.1128/AEM.64.9.3346-3351.1998
Growth rate regulation of rRNA content of a marine synechococcus (Cyanobacterium) strain
Abstract
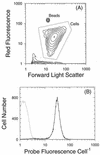
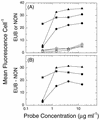
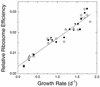
The relationship between growth rate and rRNA content in a marine Synechococcus strain was examined. A combination of flow cytometry and whole-cell hybridization with fluorescently labeled 16S rRNA-targeted oligonucleotide probes was used to measure the rRNA content of Synechococcus strain WH8101 cells grown at a range of light-limited growth rates. The sensitivity of this approach was sufficient for the analysis of rRNA even in very slowly growing Synechococcus cells (&mgr; = 0.15 day-1). The relationship between growth rate and cellular rRNA content comprised three phases: (i) at low growth rates (< approximately 0.7 day-1), rRNA cell-1 remained approximately constant; (ii) at intermediate rates ( approximately 0. 7 - 1.6 day-1), rRNA cell-1 increased proportionally with growth rate; and (iii) at the highest, light-saturated rates (> approximately 1.6 day-1), rRNA cell-1 dropped abruptly. Total cellular RNA (as measured with the nucleic acid stain SYBR Green II) was well correlated with the probe-based measure of rRNA and varied in a similar manner with growth rate. Mean cell volume and rRNA concentration (amount of rRNA per cubic micrometer) were related to growth rate in a manner similar to rRNA cell-1, although the overall magnitude of change in both cases was reduced. These patterns are hypothesized to reflect an approximately linear increase in ribosome efficiency with increasing growth rate, which is consistent with the prevailing prokaryotic model at low growth rates. Taken together, these results support the notion that measurements of cellular rRNA content might be useful for estimating in situ growth rates in natural Synechococcus populations.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

