Mercury resistance is encoded by transferable giant linear plasmids in two chesapeake bay Streptomyces strains
- PMID: 9726886
- PMCID: PMC106736
- DOI: 10.1128/AEM.64.9.3383-3388.1998
Mercury resistance is encoded by transferable giant linear plasmids in two chesapeake bay Streptomyces strains
Abstract
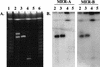
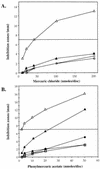
The Streptomyces strains CHR3 and CHR28, isolated from the Baltimore Inner Harbor, contained two and one, respectively, giant linear plasmids which carry terminally bound proteins. The plasmids pRJ3L (322 kb), from CHR3, and pRJ28 (330 kb), from CHR28, carry genes homologous to the previously characterized chromosomal Streptomyces lividans 66 operon encoding resistance against mercuric compounds. Both plasmids are transmissible (without any detectable rearrangement) to the chloramphenicol-resistant S. lividans TK24 strain lacking plasmids and carrying a chromosomal deletion of the mer operon. S. lividans TK24 conjugants harboring pRJ3L or pRJ28 exhibited profiles of mercury resistance to mercuric compounds similar to those of Streptomyces strains CHR3 and CHR28.
Figures

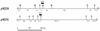

Similar articles
-
Mercury-resistant actinomycetes from the Chesapeake Bay.FEMS Microbiol Lett. 1998 May 1;162(1):177-84. doi: 10.1111/j.1574-6968.1998.tb12996.x. FEMS Microbiol Lett. 1998. PMID: 9595680
-
Interspecific transfer of Streptomyces giant linear plasmids in sterile amended soil microcosms.Appl Environ Microbiol. 2000 Feb;66(2):529-34. doi: 10.1128/AEM.66.2.529-534.2000. Appl Environ Microbiol. 2000. PMID: 10653714 Free PMC article.
-
Regulation of the operon responsible for broad-spectrum mercury resistance in Streptomyces lividans 1326.Mol Gen Genet. 1996 Jun 12;251(3):307-15. doi: 10.1007/BF02172521. Mol Gen Genet. 1996. PMID: 8676873
-
Cloning and sequence analysis of the mercury resistance operon of Streptomyces sp. Strain CHR28 reveals a novel putative second regulatory gene.J Bacteriol. 2000 Apr;182(8):2345-9. doi: 10.1128/JB.182.8.2345-2349.2000. J Bacteriol. 2000. PMID: 10735885 Free PMC article.
-
Bacterial resistances to inorganic mercury salts and organomercurials.Plasmid. 1992 Jan;27(1):4-16. doi: 10.1016/0147-619x(92)90002-r. Plasmid. 1992. PMID: 1311113 Review.
Cited by
-
-Genomic data mining of the marine actinobacteria Streptomyces sp. H-KF8 unveils insights into multi-stress related genes and metabolic pathways involved in antimicrobial synthesis.PeerJ. 2017 Feb 14;5:e2912. doi: 10.7717/peerj.2912. eCollection 2017. PeerJ. 2017. PMID: 28229018 Free PMC article.
-
Comparative Metagenomic Study of Rhizospheric and Bulk Mercury-Contaminated Soils in the Mining District of Almadén.Front Microbiol. 2022 Mar 7;13:797444. doi: 10.3389/fmicb.2022.797444. eCollection 2022. Front Microbiol. 2022. PMID: 35330761 Free PMC article.
-
Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2.J Bacteriol. 2001 Apr;183(7):2172-7. doi: 10.1128/JB.183.7.2172-2177.2001. J Bacteriol. 2001. PMID: 11244054 Free PMC article.
-
The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic island.Genome Biol Evol. 2013;5(6):1165-75. doi: 10.1093/gbe/evt082. Genome Biol Evol. 2013. PMID: 23709624 Free PMC article.
-
Extremophile Metal Resistance: Plasmid-Encoded Functions in Streptomyces mirabilis.Appl Environ Microbiol. 2022 Jun 14;88(11):e0008522. doi: 10.1128/aem.00085-22. Epub 2022 May 23. Appl Environ Microbiol. 2022. PMID: 35604229 Free PMC article.
References
-
- Benjamin M M, Datta A R. Modified pulsed field gel electrophoresis technique using Pefabloc® SC for analyzing Listeria monocytogenes DNA. Biochemica. 1995;2:30–31.
-
- Berdy J. Recent advances in and prospects of antibiotic research. Process Biochem. 1988;15:28–35.
-
- Bibb M, Ward J M, Kieser T, Cohen S N, Hopwood D A. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol Gen Genet. 1981;18:230–240. - PubMed
-
- Birren B, Lai E. Pulsed field gel electrophoresis: a practical guide. New York, N.Y: Academic Press, Inc.; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

