PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5' nuclease (TaqMan) assay
- PMID: 9726887
- PMCID: PMC106737
- DOI: 10.1128/AEM.64.9.3389-3396.1998
PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5' nuclease (TaqMan) assay
Abstract
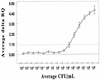
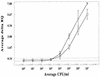
Presumptive identification of Escherichia coli O157:H7 is possible in an individual, nonmultiplexed PCR if the reaction targets the enterohemorrhagic E. coli (EHEC) eaeA gene. In this report, we describe the development and evaluation of the sensitivity and specificity of a PCR-based 5' nuclease assay for presumptively detecting E. coli O157:H7 DNA. The specificity of the eaeA-based 5' nuclease assay system was sufficient to correctly identify all E. coli O157:H7 strains evaluated, mirroring the previously described specificity of the PCR primers. The SZ-primed, eaeA-targeted 5' nuclease detection assay was capable of rapid, semiautomated, presumptive detection of E. coli O157:H7 when >/=10(3) CFU/ml was present in modified tryptic soy broth (mTSB) or modified E. coli broth and when >/=10(4) CFU/ml was present in ground beef-mTSB mixtures. Incorporating an immunomagnetic separation (IMS) step, followed by a secondary enrichment culturing step and DNA recovery with a QIAamp tissue kit (Qiagen), improved the detection threshold to >/=10(2) CFU/ml. Surprisingly, immediately after IMS, the sensitivity of culturing on sorbitol MacConkey agar containing cefeximine and tellurite (CT-SMAC) was such that identifiable colonies were demonstrated only when >/=10(4) CFU/ml was present in the sample. Several factors that might be involved in creating these false-negative CT-SMAC culture results are discussed. The SZ-primed, eaeA-targeted 5' nuclease detection system demonstrated that it can be integrated readily into standard culturing procedures and that the assay can be useful as a rapid, automatable process for the presumptive identification of E. coli O157:H7 in ground beef and potentially in other food and environmental samples.
Figures
Similar articles
-
Semi-automated fluorogenic PCR assays (TaqMan) forrapid detection of Escherichia coli O157:H7 and other shiga toxigenic E. coli.Mol Cell Probes. 1999 Aug;13(4):291-302. doi: 10.1006/mcpr.1999.0251. Mol Cell Probes. 1999. PMID: 10441202
-
PCR assay for detection of the E. coli O157:H7 eae-gene and effect of the sample preparation method on PCR detection of heat-killed E. coli O157:H7 in ground beef.Int J Food Microbiol. 1999 Nov 1;52(1-2):85-95. doi: 10.1016/s0168-1605(99)00132-4. Int J Food Microbiol. 1999. PMID: 10573395
-
Detection of enterohemorrhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and Shiga toxins.Vet Microbiol. 2003 May 29;93(3):247-60. doi: 10.1016/s0378-1135(03)00039-7. Vet Microbiol. 2003. PMID: 12695048
-
Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction.J Food Prot. 2002 Sep;65(9):1371-80. doi: 10.4315/0362-028x-65.9.1371. J Food Prot. 2002. PMID: 12233845
-
Methods for the detection and isolation of Shiga toxin-producing Escherichia coli.Symp Ser Soc Appl Microbiol. 2000;(29):133S-143S. doi: 10.1111/j.1365-2672.2000.tb05341.x. Symp Ser Soc Appl Microbiol. 2000. PMID: 10880188 Review.
Cited by
-
Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands.Appl Environ Microbiol. 2002 Oct;68(10):4853-62. doi: 10.1128/AEM.68.10.4853-4862.2002. Appl Environ Microbiol. 2002. PMID: 12324331 Free PMC article.
-
Multiplex real-time PCR method to identify Shiga toxin genes stx1 and stx2 and Escherichia coli O157:H7/H- serotype.Appl Environ Microbiol. 2003 Oct;69(10):6327-33. doi: 10.1128/AEM.69.10.6327-6333.2003. Appl Environ Microbiol. 2003. PMID: 14532101 Free PMC article.
-
Rapid 5' nuclease (TaqMan) assay for detection of virulent strains of Yersinia enterocolitica.Appl Environ Microbiol. 2000 Sep;66(9):4131-5. doi: 10.1128/AEM.66.9.4131-4135.2000. Appl Environ Microbiol. 2000. PMID: 10966441 Free PMC article.
-
Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay.J Clin Microbiol. 2000 Nov;38(11):3971-8. doi: 10.1128/JCM.38.11.3971-3978.2000. J Clin Microbiol. 2000. PMID: 11060054 Free PMC article.
-
Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7.J Clin Microbiol. 2000 May;38(5):1786-90. doi: 10.1128/JCM.38.5.1786-1790.2000. J Clin Microbiol. 2000. PMID: 10790100 Free PMC article.
References
-
- Alexander E R, Boase J, Davis M, Kirchner L, Osaki C, Tanino T, Samadpour M, Tarr P, Goldoft M, Lankford S, Kobyashi J, Stehr-Green P, Bradley P, Hinton B, Tighe P, Pearson B, Flores G R, Abbott S, Bryant R, Werner S B, Vugia D J. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California, 1994. Morbid Mortal Weekly Rep. 1995;44:157–159. - PubMed
-
- Batt C A. Molecular diagnostics for dairy-borne pathogens. J Dairy Sci. 1997;80:220–229. - PubMed
-
- Belongia E A, MacDonald K L, Parham G L, White K E, Korlath, Lobato M N, Strand S M, Casale K A, Osterholm M T. An outbreak of Escherichia coli O157:H7 colitis associated with consumption of precooked meat patties. J Infect Dis. 1991;164:338–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

