Pseudomonas sp. strain 273, an aerobic alpha, omega-dichloroalkaneDegrading bacterium
- PMID: 9726906
- PMCID: PMC106756
- DOI: 10.1128/AEM.64.9.3507-3511.1998
Pseudomonas sp. strain 273, an aerobic alpha, omega-dichloroalkaneDegrading bacterium
Abstract
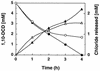
A gram-negative, aerobic bacterium was isolated from soil; this bacterium grew in 50% (vol/vol) suspensions of 1,10-dichlorodecane (1,10-DCD) as the sole source of carbon and energy. Phenotypic and small-subunit ribosomal RNA characterizations identified the organism, designated strain 273, as a member of the genus Pseudomonas. After induction with 1,10-DCD, Pseudomonas sp. strain 273 released stoichiometric amounts of chloride from C5 to C12 alpha, omega-dichloroalkanes in the presence of oxygen. No dehalogenation occurred under anaerobic conditions. The best substrates for dehalogenation and growth were C9 to C12 chloroalkanes. The isolate also grew with nonhalogenated aliphatic compounds, and decane-grown cells dechlorinated 1,10-DCD without a lag phase. In addition, cells grown on decane dechlorinated 1,10-DCD in the presence of chloramphenicol, indicating that the 1,10-DCD-dechlorinating enzyme system was also induced by decane. Other known alkane-degrading Pseudomonas species did not grow with 1,10-DCD as a carbon source. Dechlorination of 1,10-DCD was demonstrated in cell extracts of Pseudomonas sp. strain 273. Cell-free activity was strictly oxygen dependent, and NADH stimulated dechlorination, whereas EDTA had an inhibitory effect.
Figures


References
-
- Armfield S J, Sallis P J, Baker P B, Bull A T, Hardman D J. Dehalogenation of haloalkanes by Rhodococcus erythropolis Y2. Biodegradation. 1995;6:237–246. - PubMed
-
- Auling G. Pseudomonads. In: Rehm H J, Reed G, editors. Biotechnology. 2nd ed. Vol. 1. Weinheim, Germany: VCH; 1993. pp. 401–433.
-
- Behret H. Beratergremium für umweltrelevante Altstoffe. Vol. 93. Weinheim, Germany: Bundesumweltamt; 1993. Chlorparaffine; pp. 74–102.
-
- Curragh H, Flynn O, Larkin M J, Stafford T M, Hamilton J T G, Harper D B. Haloalkane degradation and assimilation by Rhodococcus rhodochrous NCIMB 13064. Microbiology. 1994;140:1433–1422. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

