Molecular and biotechnological aspects of microbial proteases
- PMID: 9729602
- PMCID: PMC98927
- DOI: 10.1128/MMBR.62.3.597-635.1998
Molecular and biotechnological aspects of microbial proteases
Abstract
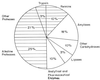
Proteases represent the class of enzymes which occupy a pivotal position with respect to their physiological roles as well as their commercial applications. They perform both degradative and synthetic functions. Since they are physiologically necessary for living organisms, proteases occur ubiquitously in a wide diversity of sources such as plants, animals, and microorganisms. Microbes are an attractive source of proteases owing to the limited space required for their cultivation and their ready susceptibility to genetic manipulation. Proteases are divided into exo- and endopeptidases based on their action at or away from the termini, respectively. They are also classified as serine proteases, aspartic proteases, cysteine proteases, and metalloproteases depending on the nature of the functional group at the active site. Proteases play a critical role in many physiological and pathophysiological processes. Based on their classification, four different types of catalytic mechanisms are operative. Proteases find extensive applications in the food and dairy industries. Alkaline proteases hold a great potential for application in the detergent and leather industries due to the increasing trend to develop environmentally friendly technologies. There is a renaissance of interest in using proteolytic enzymes as targets for developing therapeutic agents. Protease genes from several bacteria, fungi, and viruses have been cloned and sequenced with the prime aims of (i) overproduction of the enzyme by gene amplification, (ii) delineation of the role of the enzyme in pathogenecity, and (iii) alteration in enzyme properties to suit its commercial application. Protein engineering techniques have been exploited to obtain proteases which show unique specificity and/or enhanced stability at high temperature or pH or in the presence of detergents and to understand the structure-function relationships of the enzyme. Protein sequences of acidic, alkaline, and neutral proteases from diverse origins have been analyzed with the aim of studying their evolutionary relationships. Despite the extensive research on several aspects of proteases, there is a paucity of knowledge about the roles that govern the diverse specificity of these enzymes. Deciphering these secrets would enable us to exploit proteases for their applications in biotechnology.
Figures
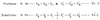
 ; S1 through Sn and S1′ through
Sn′ are the specificity subsites on the enzyme, while P1 through Pn and
P1′ through Pn′ are the residues on the substrate accommodated by the
subsites on the enzyme.
; S1 through Sn and S1′ through
Sn′ are the specificity subsites on the enzyme, while P1 through Pn and
P1′ through Pn′ are the residues on the substrate accommodated by the
subsites on the enzyme.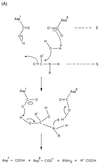















References
-
- Akimkina T V, Nosovskaya E A, Kostrov S V. Cloning and expression of the neutral proteinase gene of Bacillus cereus in Bacillus subtiliscells. Mol Biol. 1992;26:418–423. - PubMed
-
- Allen C, Stromer V K, Smith F D, Lacy G H, Mount M S. Complementation of an Erwinia carotovora subsp. carotovoraprotease mutant with a protease encoding cosmid. Mol Gen Genet. 1986;202:276–279.
-
- Amerik A, Chistyakova L G, Ostroumova N I, Gurevich A I, Antonov V K. Cloning, expression and structure of the truncated functionally active lon gene of Escherichia coli. Bioorg Khim. 1988;14:408–411. - PubMed
-
- Argos P. A sensitive procedure to compare amino acid sequences. J Mol Biol. 1987;193:385–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

