Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells
- PMID: 9730887
- PMCID: PMC2213390
- DOI: 10.1084/jem.188.5.855
Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells
Abstract
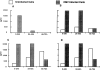
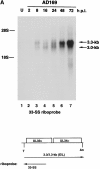
Human cytomegalovirus (HCMV), a betaherpesvirus, has developed several ways to evade the immune system, notably downregulation of cell surface expression of major histocompatibility complex class I heavy chains. Here we report that HCMV has devised another means to compromise immune surveillance mechanisms. Extracellular accumulation of both constitutively produced monocyte chemoattractant protein (MCP)-1 and tumor necrosis factor-superinduced RANTES (regulated on activation, normal T cell expressed and secreted) was downregulated in HCMV-infected fibroblasts in the absence of transcriptional repression or the expression of polyadenylated RNA for the cellular chemokine receptors CCR-1, CCR-3, and CCR-5. Competitive binding experiments demonstrated that HCMV-infected cells bind RANTES, MCP-1, macrophage inflammatory protein (MIP)-1beta, and MCP-3, but not MCP-2, to the same receptor as does MIP-1alpha, which is not expressed in uninfected cells. HCMV encodes three proteins with homology to CC chemokine receptors: US27, US28, and UL33. Cells infected with HCMV mutants deleted of US28, or both US27 and US28 genes, failed to downregulate extracellular accumulation of either RANTES or MCP-1. In contrast, cells infected with a mutant deleted of US27 continues to bind and downregulate those chemokines. Depletion of chemokines from the culture medium was at least partially due to continuous internalization of extracellular chemokine, since exogenously added, biotinylated RANTES accumulated in HCMV-infected cells. Thus, HCMV can modify the chemokine environment of infected cells through intense sequestering of CC chemokines, mediated principally by expression of the US28-encoded chemokine receptor.
Figures








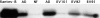
References
-
- Davis-Poynter NJ, Farrell HE. Masters of deception: a review of herpesvirus immune evasion strategies. Immunol Cell Biol. 1996;74:513–522. - PubMed
-
- Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. - PubMed
-
- Murphy PM. Molecular piracy of chemokine receptors by herpesviruses. Infect Agents Dis. 1994;3:137–154. - PubMed
-
- Smith GL. Virus proteins that bind cytokines, chemokines or interferons. Curr Opin Immunol. 1996;8:467–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

