Highly restricted T cell repertoire shaped by a single major histocompatibility complex-peptide ligand in the presence of a single rearranged T cell receptor beta chain
- PMID: 9730891
- PMCID: PMC2213398
- DOI: 10.1084/jem.188.5.897
Highly restricted T cell repertoire shaped by a single major histocompatibility complex-peptide ligand in the presence of a single rearranged T cell receptor beta chain
Abstract
The T cell repertoire is shaped by positive and negative selection of thymocytes through the interaction of alpha/beta-T cell receptors (TCR) with self-peptides bound to self-major histocompatibility complex (MHC) molecules. However, the involvement of specific TCR-peptide contacts in positive selection remains unclear. By fixing TCR-beta chains with a single rearranged TCR-beta irrelevant to the selecting ligand, we show here that T cells selected to mature on a single MHC-peptide complex express highly restricted TCR-alpha chains in terms of Valpha usage and amino acid residue of their CDR3 loops, whereas such restriction was not observed with those selected by the same MHC with diverse sets of self-peptides including this peptide. Thus, we visualized the TCR structure required to survive positive selection directed by this single ligand. Our findings provide definitive evidence that specific recognition of self-peptides by TCR could be involved in positive selection of thymocytes.
Figures

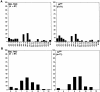

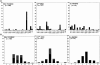

References
-
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. - PubMed
-
- Bevan MJ. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977;269:417–418. - PubMed
-
- Kisielow P, Teh HS, Blüthmann H, von Boehmer H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature. 1988;335:730–733. - PubMed
-
- Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Selective expression of an antigen receptor on CD8-bearing T lymphocytes in transgenic mice. Nature. 1988;335:271–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

