Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3'-end formation and excision of the 3'-terminal intron
- PMID: 9732273
- PMCID: PMC317118
- DOI: 10.1101/gad.12.17.2759
Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3'-end formation and excision of the 3'-terminal intron
Abstract
We have analyzed transcription termination, 3'-end formation, and excision of the 3'-terminal intron in vivo in the Balbiani ring 1 (BR1) gene and its pre-mRNA. We show that full-length RNA transcripts are evenly spaced on the gene from a position 300 bp upstream to a region 500-700 bp downstream of the polyadenylation sequence. Very few full-length transcripts and no short, cleaved, nascent transcripts could be observed downstream of this region. Pre-mRNA with 10-20 adenylate residues accumulates at the active gene and then rapidly leaves from the gene locus. Only polyadenylated pre-mRNAs could be detected in the nucleoplasm. Our results are consistent with the hypothesis that transcription termination occurs in a narrow region for the majority of transcripts, simultaneous with 3'-end formation. Excision of the 3'-terminal intron occurs before 3'-end formation in about 5% of the nascent transcripts. When transcription terminates, 3' cleavage takes place and 10-20 adenylate residues are added, the 3'-terminal intron is excised from additionally about 75% of the pre-mRNA at the gene locus. Our data support a close temporal and spatial coupling of transcription termination and the cleavage and initial polyadenylation of 3'-end formation. Excision of the 3'-terminal intron is highly stimulated as the cleavage/polyadenylation complex assembles and 3'-end formation is initiated.
Figures

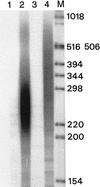




References
-
- Baurén G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - PubMed
-
- Belikov S, Paulsson G, Wieslander L. Promoter regions of four Balbiani ring genes exhibit a common salivary gland specific chromatin organisation which is independent of the rate of transcription. Mol & Gen Genet. 1998;258:420–426. - PubMed
-
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA mutation in yeast. Science. 1998;280:298–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
