The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley
- PMID: 9733524
- PMCID: PMC34872
- DOI: 10.1104/pp.118.1.37
The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley
Abstract
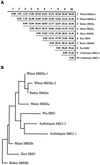
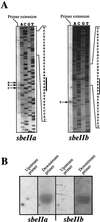
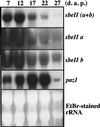
The sbeIIa and sbeIIb genes, encoding starch-branching enzyme (SBE) IIa and SBEIIb in barley (Hordeum vulgare L.), have been isolated. The 5' portions of the two genes are strongly divergent, primarily due to the 2064-nucleotide-long intron 2 in sbeIIb. The sequence of this intron shows that it contains a retro-transposon-like element. Expression of sbeIIb but not sbeIIa was found to be endosperm specific. The temporal expression patterns for sbeIIa and sbeIIb were similar and peaked around 12 d after pollination. DNA gel-blot analysis demonstrated that sbeIIa and sbeIIb are both single-copy genes in the barley genome. By fluorescence in situ hybridization, the sbeIIa and sbeIIb genes were mapped to chromosomes 2 and 5, respectively. The cDNA clones for SBEIIa and SBEIIb were isolated and sequenced. The amino acid sequences of SBEIIa and SBEIIb were almost 80% identical. The major structural difference between the two enzymes was the presence of a 94-amino acid N-terminal extension in the SBEIIb precursor. The (beta/alpha)8-barrel topology of the alpha-amylase superfamily and the catalytic residues implicated in branching enzymes are conserved in both barley enzymes.
Figures






References
-
- Baba T, Kimura K, Mizuno K, Etoh H, Ishida Y, Shida O, Arai Y. Sequence conservation of the catalytic regions of amylolytic enzymes in maize branching enzyme-I. Biochem Biophys Res Commun. 1991;181:87–94. - PubMed
-
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. - PubMed
-
- Bennetzen JL. The contributions of retroelements to plant genome organization, function and evolution. Trends Microbiol. 1996;4:347–353. - PubMed
-
- Bhattacharyya MK, Smith AM, Ellis THN, Hedley C, Martin C. The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch branching enzyme. Cell. 1990;60:115–122. - PubMed
-
- Boyer CD, Preiss J. Multiple forms of (1→4)-α-d-glucan, (1→4)-α-glucan-6-glycosyl transferse from developing Zea mays L. kernels. Carbohydr Res. 1978;61:321–334.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

