Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase
- PMID: 9733547
- PMCID: PMC34866
- DOI: 10.1104/pp.118.1.275
Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase
Abstract
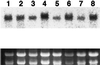
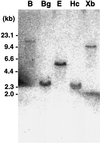
Phosphoribosyl-ATP pyrophosphohydrolase (PRA-PH) and phosphoribosyl-AMP cyclohydrolase (PRA-CH) are encoded by HIS4 in yeast and by hisIE in bacteria and catalyze the second and the third step, respectively, in the histidine biosynthetic pathway. By complementing a hisI mutation of Escherichia coli with an Arabidopsis cDNA library, we isolated an Arabidopsis cDNA (At-IE) that possesses these two enzyme activities. The At-IE cDNA encodes a bifunctional protein of 281 amino acids with a calculated molecular mass of 31,666 D. Genomic DNA-blot analysis with the At-IE cDNA as a probe revealed a single-copy gene in Arabidopsis, and RNA-blot analysis showed that the At-IE gene was expressed ubiquitously throughout development. Sequence comparison suggested that the At-IE protein has an N-terminal extension of about 50 amino acids with the properties of a chloroplast transit peptide. We demonstrated through heterologous expression studies in E. coli that the functional domains for the PRA-CH (hisI) and PRA-PH (hisE) resided in the N-terminal and the C-terminal halves, respectively, of the At-IE protein.
Figures






Similar articles
-
Molecular cloning and characterization of ATP-phosphoribosyl transferase from Arabidopsis, a key enzyme in the histidine biosynthetic pathway.Plant Physiol. 2000 Mar;122(3):907-14. doi: 10.1104/pp.122.3.907. Plant Physiol. 2000. PMID: 10712555 Free PMC article.
-
Molecular cloning and characterization of the gene encoding N'-[(5'-phosphoribosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (BBM II) isomerase from Arabidopsis thaliana.Mol Gen Genet. 1998 Aug;259(2):216-23. doi: 10.1007/s004380050807. Mol Gen Genet. 1998. PMID: 9747713
-
Structural and mechanistic insights into the bifunctional HISN2 enzyme catalyzing the second and third steps of histidine biosynthesis in plants.Sci Rep. 2021 May 6;11(1):9647. doi: 10.1038/s41598-021-88920-2. Sci Rep. 2021. PMID: 33958623 Free PMC article.
-
Exploring the role of the histidine biosynthetic hisF gene in cellular metabolism and in the evolution of (ancestral) genes: from LUCA to the extant (micro)organisms.Microbiol Res. 2020 Nov;240:126555. doi: 10.1016/j.micres.2020.126555. Epub 2020 Jul 9. Microbiol Res. 2020. PMID: 32673985 Review.
-
Histidine biosynthetic pathway and genes: structure, regulation, and evolution.Microbiol Rev. 1996 Mar;60(1):44-69. doi: 10.1128/mr.60.1.44-69.1996. Microbiol Rev. 1996. PMID: 8852895 Free PMC article. Review. No abstract available.
Cited by
-
Catalytic zinc site and mechanism of the metalloenzyme PR-AMP cyclohydrolase.Biochemistry. 2012 Jul 24;51(29):5791-803. doi: 10.1021/bi300391m. Epub 2012 Jul 9. Biochemistry. 2012. PMID: 22741521 Free PMC article.
-
Comparative proteomic analysis reveals the regulatory network of the veA gene during asexual and sexual spore development of Aspergillus cristatus.Biosci Rep. 2018 Jul 31;38(4):BSR20180067. doi: 10.1042/BSR20180067. Print 2018 Aug 31. Biosci Rep. 2018. Retraction in: Biosci Rep. 2021 Apr 30;41(4):BSR-20180067_RET. doi: 10.1042/BSR-20180067_RET. PMID: 29773679 Free PMC article. Retracted.
-
Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants.Plant Cell. 2005 Jul;17(7):2089-106. doi: 10.1105/tpc.104.030577. Epub 2005 May 27. Plant Cell. 2005. PMID: 15923352 Free PMC article.
-
Molecular cloning and characterization of ATP-phosphoribosyl transferase from Arabidopsis, a key enzyme in the histidine biosynthetic pathway.Plant Physiol. 2000 Mar;122(3):907-14. doi: 10.1104/pp.122.3.907. Plant Physiol. 2000. PMID: 10712555 Free PMC article.
-
Identification and molecular characterization of mitochondrial ferredoxins and ferredoxin reductase from Arabidopsis.Plant Mol Biol. 2003 Jul;52(4):817-30. doi: 10.1023/a:1025015811141. Plant Mol Biol. 2003. PMID: 13677469
References
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

