Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating
- PMID: 9733692
- PMCID: PMC107514
- DOI: 10.1128/JB.180.18.4886-4892.1998
Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating
Abstract
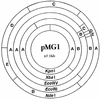
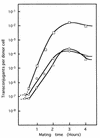
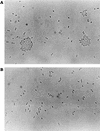
Plasmid pMG1 (65.1 kb) was isolated from a gentamicin-resistant Enterococcus faecium clinical isolate and was found to encode gentamicin resistance. EcoRI restriction of pMG1 produced five fragments, A through E, with molecular sizes of 50.2, 11.5, 2.0, 0.7, and 0.7 kb, respectively. The clockwise order of the fragments was ACDEB. pMG1 transferred at high frequency to Enterococcus strains in broth mating. pMG1 transferred between Enterococcus faecalis strains, between E. faecium strains, and between E. faecium and E. faecalis strains at a frequency of approximately 10(-4) per donor cell after 3 h of mating. The pMG1 transfers were not induced by the exposure of the donor cell to culture filtrates of plasmid-free E. faecalis FA2-2 or an E. faecium strain. Mating aggregates were not observed by the naked eye during broth mating. Small mating aggregates of several cells in the broth matings were observed by microscopy, while no aggregates of donor cells which had been exposed to a culture filtrate of E. faecalis FA2-2 or an E. faecium strain were observed, even by microscopy. pMG1 DNA did not show any homology in Southern hybridization with that of the pheromone-responsive plasmids and broad-host-range plasmids pAMbeta1 and pIP501. These results indicate that there is another efficient transfer system in the conjugative plasmids of Enterococcus and that this system is different from the pheromone-induced transfer system of E. faecalis plasmids.
Figures

References
-
- Behnke D, Gilmore M S. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184:115–120. - PubMed
-
- Christie P J, Dunny G M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986;15:230–241. - PubMed
-
- Clewell D B. Bacterial sex pheromone-induced plasmid transfer. Cell. 1993;73:9–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

