Two functionally distinct regions upstream of the cbbI operon of Rhodobacter sphaeroides regulate gene expression
- PMID: 9733694
- PMCID: PMC107516
- DOI: 10.1128/JB.180.18.4903-4911.1998
Two functionally distinct regions upstream of the cbbI operon of Rhodobacter sphaeroides regulate gene expression
Abstract
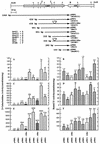
A number of cbbFI::lacZ translational fusion plasmids containing various lengths of sequence 5' to the form I (cbbI) Calvin-Benson-Bassham cycle operon (cbbFIcbbPIcbbAIcbbLIcbbSI) of Rhodobacter sphaeroides were constructed. Expression of beta-galactosidase was monitored under a variety of growth conditions. It was found that 103 bp of sequence upstream of the cbbFI transcription start was sufficient to confer low levels of regulated cbbI promoter expression; this activity was dependent on the presence of an intact cbbR gene. Additionally, R. sphaeroides CbbR was shown to bind to the region between 9 and 100 bp 5' to the cbbFI transcription start. Inclusion of an additional upstream sequence, from 280 to 636 bp 5' to cbbFI, resulted in a significant increase in regulated cbbI promoter expression under all growth conditions tested. A 50-bp region responsible for the majority of this increase occurs between 280 and 330 bp 5' to cbbFI. The additional 306 bp of upstream sequence from 330 to 636 bp also appears to play a positive regulatory role. A 4-bp deletion 281 to 284 bp 5' to cbbFI significantly reduced cbbI expression while the proper regulatory pattern was retained. These studies provide evidence for the presence of two functionally distinct regions of the cbbI promoter, with the distal domain providing significant regulated promoter activity that adheres to the normal pattern of expression.
Figures



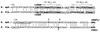
Similar articles
-
Interactions of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides.J Biol Chem. 2003 May 2;278(18):16443-50. doi: 10.1074/jbc.M211267200. Epub 2003 Feb 24. J Biol Chem. 2003. PMID: 12601011
-
Positive and negative regulation of sequences upstream of the form II cbb CO2 fixation operon of Rhodobacter sphaeroides.J Bacteriol. 1994 Dec;176(23):7299-308. doi: 10.1128/jb.176.23.7299-7308.1994. J Bacteriol. 1994. PMID: 7961502 Free PMC article.
-
Nucleotide sequence and functional analysis of cbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides.J Bacteriol. 1993 Sep;175(18):5778-84. doi: 10.1128/jb.175.18.5778-5784.1993. J Bacteriol. 1993. PMID: 8376325 Free PMC article.
-
Genetics and control of CO(2) assimilation in the chemoautotroph Ralstonia eutropha.Arch Microbiol. 2002 Aug;178(2):85-93. doi: 10.1007/s00203-002-0441-3. Epub 2002 Jun 14. Arch Microbiol. 2002. PMID: 12115053 Review.
-
Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria.FEMS Microbiol Rev. 1997 Sep;21(2):135-55. doi: 10.1111/j.1574-6976.1997.tb00348.x. FEMS Microbiol Rev. 1997. PMID: 9348665 Review.
Cited by
-
Differential accumulation of form I RubisCO in Rhodopseudomonas palustris CGA010 under Photoheterotrophic growth conditions with reduced carbon sources.J Bacteriol. 2009 Jul;191(13):4243-50. doi: 10.1128/JB.01795-08. Epub 2009 Apr 17. J Bacteriol. 2009. PMID: 19376869 Free PMC article.
-
Regulatory twist and synergistic role of metabolic coinducer- and response regulator-mediated CbbR-cbbI interactions in Rhodopseudomonas palustris CGA010.J Bacteriol. 2013 Apr;195(7):1381-8. doi: 10.1128/JB.02060-12. Epub 2013 Jan 4. J Bacteriol. 2013. PMID: 23292778 Free PMC article.
-
Amino acid residues of RegA important for interactions with the CbbR-DNA complex of Rhodobacter sphaeroides.J Bacteriol. 2014 Sep;196(17):3179-90. doi: 10.1128/JB.01842-14. Epub 2014 Jun 23. J Bacteriol. 2014. PMID: 24957624 Free PMC article.
-
Phosphoribulokinase mediates nitrogenase-induced carbon dioxide fixation gene repression in Rhodobacter sphaeroides.Microbiology (Reading). 2015 Nov;161(11):2184-91. doi: 10.1099/mic.0.000160. Epub 2015 Aug 24. Microbiology (Reading). 2015. PMID: 26306848 Free PMC article.
-
Differential expression of the CO2 fixation operons of Rhodobacter sphaeroides by the Prr/Reg two-component system during chemoautotrophic growth.J Bacteriol. 2002 Dec;184(23):6654-64. doi: 10.1128/JB.184.23.6654-6664.2002. J Bacteriol. 2002. PMID: 12426354 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley and Sons, Inc.; 1987.
-
- Bishop R E, Weiner J H. Overproduction, solubilization, purification and DNA-binding properties of AmpR from Citrobacter freundii. Eur J Biochem. 1993;213:405–412. - PubMed
-
- Cantrell A, Bryant D A. Molecular cloning and nucleotide sequence of the psaA and psaB genes of the cyanobacterium Synechococcus sp. PCC 7002. Plant Mol Biol. 1987;9:453–468. - PubMed
-
- Chen J-H, Gibson J L, Macue L A, Tabita F R. Identification, expression, and deduced primary structure of transketolase and other enzymes encoded within the form II CO2 fixation operon of Rhodobacter sphaeroides. J Biol Chem. 1991;266:20447–20452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

