Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin
- PMID: 9733697
- PMCID: PMC107519
- DOI: 10.1128/JB.180.18.4929-4937.1998
Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin
Abstract
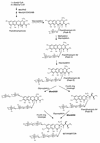
Mithramycin is an antitumor polyketide drug produced by Streptomyces argillaceus that contains two deoxysugar chains, a disaccharide consisting of two D-olivoses and a trisaccharide consisting of a D-olivose, a D-oliose, and a D-mycarose. From a cosmid clone (cosAR3) which confers resistance to mithramycin in streptomycetes, a 3-kb PstI-XhoI fragment was sequenced, and two divergent genes (mtmGI and mtmGII) were identified. Comparison of the deduced products of both genes with proteins in databases showed similarities with glycosyltransferases and glucuronosyltransferases from different sources, including several glycosyltransferases involved in sugar transfer during antibiotic biosynthesis. Both genes were independently inactivated by gene replacement, and the mutants generated (M3G1 and M3G2) did not produce mithramycin. High-performance liquid chromatography analysis of ethyl acetate extracts of culture supernatants of both mutants showed the presence of several peaks with the characteristic spectra of mithramycin biosynthetic intermediates. Four compounds were isolated from both mutants by preparative high-performance liquid chromatography, and their structures were elucidated by physicochemical methods. The structures of these compounds were identical in both mutants, and the compounds are suggested to be glycosylated intermediates of mithramycin biosynthesis with different numbers of sugar moieties attached to C-12a-O of a tetracyclic mithramycin precursor and to C-2-O of mithramycinone: three tetracyclic intermediates containing one sugar (premithramycin A1), two sugars (premithramycin A2), or three sugars (premithramycin A3) and one tricyclic intermediate containing a trisaccharide chain (premithramycin A4). It is proposed that the glycosyltransferases encoded by mtmGI and mtmGII are responsible for forming and transferring the disaccharide during mithramycin biosynthesis. From the structures of the new metabolites, a new biosynthetic sequence regarding late steps of mithramycin biosynthesis can be suggested, a sequence which includes glycosyl transfer steps prior to the final shaping of the aglycone moiety of mithramycin.
Figures




References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bechthold A, Sohng J K, Smith T M, Chu X, Floss H G. Identification of Streptomyces violaceoruber Tü22 genes involved in the biosynthesis of granaticin. Mol Gen Genet. 1995;248:610–620. - PubMed
-
- Berlin Y A, Kolosov M N, Vasina I V, Yartseva I V. The structure of chromocyclomycin. J Chem Soc Chem Commun. 1968;1968:762–763.
-
- Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. - PubMed
-
- Blanco G, Fu H, Méndez C, Khosla C, Salas J A. Deciphering the biosynthetic origin of the aglycone of the aureolic acid group of anti-tumor agents. Chem Biol. 1996;3:193–196. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

