A region in Bacillus subtilis sigmaH required for Spo0A-dependent promoter activity
- PMID: 9733708
- PMCID: PMC107530
- DOI: 10.1128/JB.180.18.4987-4990.1998
A region in Bacillus subtilis sigmaH required for Spo0A-dependent promoter activity
Abstract
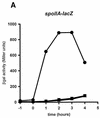
Spo0A activates transcription in Bacillus subtilis from promoters that are used by two types of RNA polymerase, RNA polymerase containing the primary sigma factor, sigmaA, and RNA polymerase containing a secondary sigma factor, known as sigmaH. The region of sigmaA near positions 356 to 359 is required for Spo0A-dependent promoter activation, possibly because Spo0A interacts with this region of sigmaA at these promoters. To determine if the amino acids in the corresponding region of sigmaH are also important in Spo0A-dependent promoter activation, we examined the effects of single alanine substitutions at 10 positions in sigmaH (201 to 210). Two alanine substitutions in sigmaH, at glutamine 201 (Q201A) and at arginine 205 (R205A), significantly decreased activity from the Spo0A-dependent, sigmaH-dependent promoter spoIIA but did not affect expression from the sigmaH-dependent, Spo0A-independent promoters citGp2 and spoVG. Therefore, promoter activation by Spo0A requires homologous regions in sigmaA and sigmaH. A mutant form of Spo0A, S231F, that suppresses the sporulation defect caused by several amino acid substitutions in sigmaA did not suppress the sporulation defects caused by the Q201A and R205A substitutions in sigmaH. This result and others indicate that different surfaces of Spo0A probably interact with sigmaA and sigmaH RNA polymerases.
Figures



References
-
- Baldus J M, Buckner C M, Moran C P., Jr Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995;17:281–290. - PubMed
-
- Cormack B. Mutagenesis of cloned DNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. pp. 857–859.
-
- Feavers I M, Price V, Moir A. The regulation of the fumarase (citG) gene of Bacillus subtilis 168. Mol Gen Genet. 1988;211:465–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

