Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly
- PMID: 9733825
- PMCID: PMC110113
- DOI: 10.1128/JVI.72.10.7885-7894.1998
Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly
Abstract
Expression studies have shown that the coronavirus small envelope protein E and the much more abundant membrane glycoprotein M are both necessary and sufficient for the assembly of virus-like particles in cells. As a step toward understanding the function of the mouse hepatitis virus (MHV) E protein, we carried out clustered charged-to-alanine mutagenesis on the E gene and incorporated the resulting mutations into the MHV genome by targeted recombination. Of the four possible clustered charged-to-alanine E gene mutants, one was apparently lethal and one had a wild-type phenotype. The two other mutants were partially temperature sensitive, forming small plaques at the nonpermissive temperature. Revertant analyses of these two mutants demonstrated that the created mutations were responsible for the temperature-sensitive phenotype of each and provided support for possible interactions among E protein monomers. Both temperature-sensitive mutants were also found to be markedly thermolabile when grown at the permissive temperature, suggesting that there was a flaw in their assembly. Most significantly, when virions of one of the mutants were examined by electron microscopy, they were found to have strikingly aberrant morphology in comparison to the wild type: most mutant virions had pinched and elongated shapes that were rarely seen among wild-type virions. These results demonstrate an important, probably essential, role for the E protein in coronavirus morphogenesis.
Figures
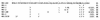





References
-
- Bennett W F, Paoni N F, Keyt B A, Botstein D, Jones A J, Presta L, Wurm F M, Zoller M J. High resolution analysis of functional determinants on human tissue-type plasminogen activator. J Biol Chem. 1991;266:5191–5201. - PubMed
-
- Boursnell M E G, Binns M M, Brown T D K. Sequencing of coronavirus IBV genomic RNA: three open reading frames in the 5′ ‘unique’ region of mRNA D. J Gen Virol. 1985;66:2253–2258. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

