Cleavage susceptibility of reovirus attachment protein sigma1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the sigma1 neck
- PMID: 9733863
- PMCID: PMC110170
- DOI: 10.1128/JVI.72.10.8205-8213.1998
Cleavage susceptibility of reovirus attachment protein sigma1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the sigma1 neck
Abstract
A requisite step in reovirus infection of the murine intestine is proteolysis of outer-capsid proteins to yield infectious subvirion particles (ISVPs). When converted to ISVPs by intestinal proteases, virions of reovirus strain type 3 Dearing (T3D) lose 90% of their original infectivity due to cleavage of viral attachment protein sigma1. In an analysis of eight field isolate strains of type 3 reovirus, we identified one additional strain, type 3 clone 31 (T3C31), that loses infectivity and undergoes sigma1 cleavage upon conversion of virions to ISVPs. We examined the sigma1 deduced amino acid sequences of T3D and the eight field isolate strains for a correlation between sequence variability and sigma1 cleavage. The sigma1 proteins of T3D and T3C31 contain a threonine at amino acid position 249, whereas an isoleucine occurs at this position in the sigma1 proteins of the remaining strains. Thr249 occupies the d position of a heptad repeat motif predicted to stabilize sigma1 oligomers through alpha-helical coiled-coil interactions. This region of sequence comprises a portion of the fibrous tail domain of sigma1 known as the neck. Substitution of Thr249 with isoleucine or leucine resulted in resistance to cleavage by trypsin, whereas replacement with asparagine did not affect cleavage susceptibility. These results demonstrate that amino acid position 249 is an independent determinant of T3D sigma1 cleavage susceptibility and that an intact heptad repeat is required to confer cleavage resistance. We performed amino-terminal sequence analysis on the sigma1 cleavage product released during trypsin treatment of T3D virions to generate ISVPs and found that trypsin cleaves sigma1 after Arg245. Thus, the sequence polymorphism at position 249 controls cleavage at a nearby site in the neck region. The relevance of these results to reovirus infection in vivo was assessed by treating virions with the contents of a murine intestinal wash under conditions that result in generation of ISVPs. The pattern of sigma1 cleavage susceptibility generated by using purified protease was reproduced in assays using the intestinal wash. These results provide a mechanistic explanation for sigma1 cleavage during exposure of virions to intestinal proteases and may account for certain strain-dependent patterns of reovirus pathogenesis.
Figures


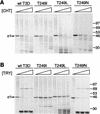
 , 0 to 67.5 μg of chymotrypsin per ml. (B) MAb G5-conjugated Sepharose containing expressed ς1 protein was treated with various concentrations of trypsin (TRY) at 4°C for 60 min. Treatment mixtures were processed as described for panel A.
, 0 to 67.5 μg of chymotrypsin per ml. (B) MAb G5-conjugated Sepharose containing expressed ς1 protein was treated with various concentrations of trypsin (TRY) at 4°C for 60 min. Treatment mixtures were processed as described for panel A.  , 0 to 18 μg of trypsin per ml.
, 0 to 18 μg of trypsin per ml.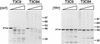
 , 0 to 67.5 μg of chymotrypsin per ml or 0 to 18 μg of trypsin per ml.
, 0 to 67.5 μg of chymotrypsin per ml or 0 to 18 μg of trypsin per ml.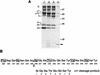
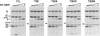
 , 0 to 83% (vol/vol) intestinal wash.
, 0 to 83% (vol/vol) intestinal wash.Similar articles
-
Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved sigma 1 protein.J Virol. 1995 Aug;69(8):5057-67. doi: 10.1128/JVI.69.8.5057-5067.1995. J Virol. 1995. PMID: 7609075 Free PMC article.
-
A glycosyl hydrolase activity of mammalian reovirus sigma1 protein can contribute to viral infection through a mucus layer.J Mol Biol. 1999 Feb 26;286(3):759-73. doi: 10.1006/jmbi.1998.2495. J Mol Biol. 1999. PMID: 10024449
-
Structural and Functional Features of the Reovirus σ1 Tail.J Virol. 2018 Jun 29;92(14):e00336-18. doi: 10.1128/JVI.00336-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29695426 Free PMC article.
-
Protein Mismatches Caused by Reassortment Influence Functions of the Reovirus Capsid.J Virol. 2018 Sep 26;92(20):e00858-18. doi: 10.1128/JVI.00858-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068646 Free PMC article.
-
Attachment and cell entry of mammalian orthoreovirus.Curr Top Microbiol Immunol. 2006;309:1-38. doi: 10.1007/3-540-30773-7_1. Curr Top Microbiol Immunol. 2006. PMID: 16909895 Review.
Cited by
-
Apoptosis induction influences reovirus replication and virulence in newborn mice.J Virol. 2013 Dec;87(23):12980-9. doi: 10.1128/JVI.01931-13. Epub 2013 Sep 25. J Virol. 2013. PMID: 24067960 Free PMC article.
-
The oncolytic effect in vivo of reovirus on tumour cells that have survived reovirus cell killing in vitro.Br J Cancer. 2006 Oct 23;95(8):1020-7. doi: 10.1038/sj.bjc.6603363. Epub 2006 Oct 3. Br J Cancer. 2006. PMID: 17047650 Free PMC article.
-
Crystal structure of reovirus attachment protein σ1 in complex with sialylated oligosaccharides.PLoS Pathog. 2011 Aug;7(8):e1002166. doi: 10.1371/journal.ppat.1002166. Epub 2011 Aug 4. PLoS Pathog. 2011. PMID: 21829363 Free PMC article.
-
Breast Tumor-Associated Metalloproteases Restrict Reovirus Oncolysis by Cleaving the σ1 Cell Attachment Protein and Can Be Overcome by Mutation of σ1.J Virol. 2019 Oct 29;93(22):e01380-19. doi: 10.1128/JVI.01380-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31462562 Free PMC article.
-
Unexpected Genetic Diversity of Two Novel Swine MRVs in Italy.Viruses. 2020 May 22;12(5):574. doi: 10.3390/v12050574. Viruses. 2020. PMID: 32456089 Free PMC article.
References
-
- Armstrong G D, Paul R W, Lee P W. Studies on reovirus receptors of L cells: virus binding characteristics and comparison with reovirus receptors of erythrocytes. Virology. 1984;138:37–48. - PubMed
-
- Banerjea A C, Brechling K A, Ray C A, Erikson H, Pickup D J, Joklik W K. High-level synthesis of biologically active reovirus protein ς1 in a mammalian expression vector system. Virology. 1988;167:601–612. - PubMed
-
- Barton, E. S., and T. S. Dermody. Unpublished observation.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

