A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus
- PMID: 9733875
- PMCID: PMC110196
- DOI: 10.1128/JVI.72.10.8309-8315.1998
A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus
Abstract
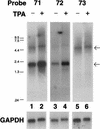
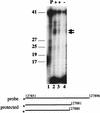
Infection with Kaposi's sarcoma-associated herpesvirus (KSHV) is closely associated with Kaposi's sarcoma (KS) and primary effusion lymphoma, with viral genomes present in a latent state in the majority of tumor cells. Here we describe a cluster of latently expressed viral genes whose mRNAs are generated from a common promoter. Two mRNAs in this region encode the latency-associated nuclear antigen, the product of open reading frame 73 (ORF73). The larger RNA, of 5.8 kb, is an unspliced transcript that includes ORF72 and -71 at its 3' end; it initiates at nucleotides (nt) 127880 to 127886 from a promoter lacking recognizable TATA elements. A less abundant mRNA, of 5.4 kb, is a variant of this transcript, in which 336 nt of 5' noncoding information has been removed by RNA splicing. A third, more abundant RNA is generated from the same promoter region via splicing from the common splice donor at nt 127813 to an acceptor 5' to ORF72; this transcript is the presumed mRNA for ORF72, which encodes the viral cyclin D homolog. All three RNAs are 3' coterminal. In situ hybridization analysis with probes that can detect all three transcripts shows that the RNAs are detectable in a large fraction of BCBL-1 cells prior to lytic induction and in >70% of KS spindle cells in primary KS tumors. This confirms that these transcripts are indeed latent RNAs and suggests a role for their products in viral persistence and/or KSHV-associated proliferation.
Figures





References
-
- Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. - PubMed
-
- Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. - PubMed
-
- Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden K, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. . (Letter.) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

