Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans
- PMID: 9733894
- PMCID: PMC110230
- DOI: 10.1128/JVI.72.10.8408-8412.1998
Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans
Abstract
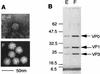
The complete nucleotide sequence of a novel enteric virus, Aichi virus, associated with nonbacterial acute gastroenteritis in humans was determined. The Aichi virus genome proved to be a single-stranded positive-sense RNA molecule with 8,251 bases excluding a poly(A) tail; it contains a large open reading frame with 7,302 nucleotides that encodes a potential polyprotein precursor of 2,433 amino acids. The genome contains a 5' nontranslated region (NTR) with 712 bases and a 3' NTR with 240 bases followed by a poly(A) tail. The structure of the genome, VPg-5' NTR-leader protein-structural proteins-nonstructural proteins-3' NTR-poly(A), was found to be typical of a picornavirus. The VP0-VP3 and VP3-VP1 cleavage sites were determined to be Q-H and Q-T, respectively, by N-terminal amino acid sequence analyses using purified virion proteins. Possible cleavage sites, Q-G, Q-A, and Q-S, which cleave P2 and P3 polyproteins were found to be similar to those of picornaviruses. A dendrogram based on 3Dpol proteins indicated that Aichi virus is genetically distinct from the known six genera of picornaviruses including entero-, rhino-, cardio-, aphtho-, and hepatovirus and echovirus 22. Considering this together with other properties of the virus (T. Yamashita, S. Kobayashi, K. Sakae, S. Nakata, S. Chiba, Y. Ishihara, and S. Isomura, J. Infect. Dis. 164:954-957, 1991), we propose that Aichi virus be regarded as a new genus of the family Picornaviridae.
Figures


References
-
- Ando T, Mulders M N, Lewis D C, Estes M K, Monroe S S, Glass R I. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch Virol. 1994;135:217–226. - PubMed
-
- Auvinen P, Hyypiä T. Echoviruses include genetically distinct serotypes. J Gen Virol. 1990;71:2133–2139. - PubMed
-
- Blacklow N R. Medical virology of small round gastroenteritis viruses. In: de la Maza L M, Peterson E M, editors. Medical virology, no. 9. New York, N.Y: Plenum Press; 1990. pp. 111–128.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

