The major open reading frame of the beta2.7 transcript of human cytomegalovirus: in vitro expression of a protein posttranscriptionally regulated by the 5' region
- PMID: 9733897
- PMCID: PMC110236
- DOI: 10.1128/JVI.72.10.8425-8429.1998
The major open reading frame of the beta2.7 transcript of human cytomegalovirus: in vitro expression of a protein posttranscriptionally regulated by the 5' region
Abstract
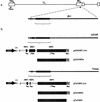
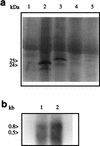
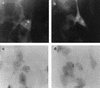
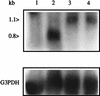
beta2.7 is the major early transcript produced during human cytomegalovirus infection. This abundantly expressed RNA is polysome associated, but no protein product has ever been detected. In this study, a stable peptide of 24 kDa was produced in vitro from the major open reading frame (ORF), TRL4. Following transient transfection, the intracellular localization was nucleolar and the expression was posttranscriptionally inhibited by the 5' sequence of the transcript, which harbors two short upstream ORFs.
Figures
References
-
- Biegalke B J, Geballe A P. Translational inhibition by cytomegalovirus transcript leader. Virology. 1990;177:657–667. - PubMed
-
- Burd C G, Dreyfuss G. Conserved structure and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. - PubMed
-
- Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

