Salmonella enteritidis: AmpC plasmid-mediated inducible beta-lactamase (DHA-1) with an ampR gene from Morganella morganii
- PMID: 9736562
- PMCID: PMC105832
- DOI: 10.1128/AAC.42.9.2352
Salmonella enteritidis: AmpC plasmid-mediated inducible beta-lactamase (DHA-1) with an ampR gene from Morganella morganii
Abstract
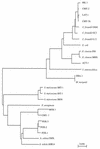
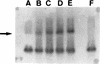
DHA-1, a plasmid-mediated cephalosporinase from a single clinical Salmonella enteritidis isolate, conferred resistance to oxyimino-cephalosporins (cefotaxime and ceftazidime) and cephamycins (cefoxitin and moxalactam), and this resistance was transferable to Escherichia coli HB101. An antagonism was observed between cefoxitin and aztreonam by the diffusion method. Transformation of the transconjugant E. coli strain with plasmid pNH5 carrying the ampD gene (whose product decreases the level of expression of ampC) resulted in an eightfold decrease in the MIC of cefoxitin. A clone with the same AmpC susceptibility pattern with antagonism was obtained, clone E. coli JM101(pSAL2-ind), and its nucleotide sequence was determined. It contained an open reading frame with 98. 7% DNA sequence identity with the ampC gene of Morganella morganii. DNA sequence analysis also identified a gene upstream of ampC whose sequence was 97% identical to the partial sequence of the ampR gene (435 bp) from M. morganii. The gene encoded a protein with an amino-terminal DNA-binding domain typical of transcriptional activators of the LysR family. Moreover, the intercistronic region between the ampC and ampR genes was 98% identical to the corresponding region from M. morganii DNA. AmpR was shown to be functional by enzyme induction and a gel mobility-shift assay. An ampG gene was also detected in a Southern blot of DNA from the S. enteritidis isolate. These findings suggest that this inducible plasmid-mediated AmpC type beta-lactamase, DHA-1, probably originated from M. morganii.
Figures



References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. Mol Biol. 1990;215:403–410. - PubMed
-
- Barnaud G, Arlet G, Danglot C, Philippon A. Cloning and sequencing of the gene encoding the AmpC β-lactamase of Morganella morganii. FEMS Microbiol Lett. 1997;148:15–20. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

