A statistical paradigm for neural spike train decoding applied to position prediction from ensemble firing patterns of rat hippocampal place cells
- PMID: 9736661
- PMCID: PMC6793233
- DOI: 10.1523/JNEUROSCI.18-18-07411.1998
A statistical paradigm for neural spike train decoding applied to position prediction from ensemble firing patterns of rat hippocampal place cells
Abstract
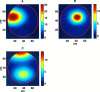
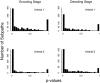
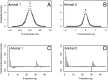
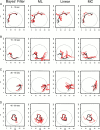
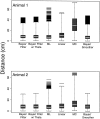
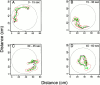
The problem of predicting the position of a freely foraging rat based on the ensemble firing patterns of place cells recorded from the CA1 region of its hippocampus is used to develop a two-stage statistical paradigm for neural spike train decoding. In the first, or encoding stage, place cell spiking activity is modeled as an inhomogeneous Poisson process whose instantaneous rate is a function of the animal's position in space and phase of its theta rhythm. The animal's path is modeled as a Gaussian random walk. In the second, or decoding stage, a Bayesian statistical paradigm is used to derive a nonlinear recursive causal filter algorithm for predicting the position of the animal from the place cell ensemble firing patterns. The algebra of the decoding algorithm defines an explicit map of the discrete spike trains into the position prediction. The confidence regions for the position predictions quantify spike train information in terms of the most probable locations of the animal given the ensemble firing pattern. Under our inhomogeneous Poisson model position was a three to five times stronger modulator of the place cell spiking activity than theta phase in an open circular environment. For animal 1 (2) the median decoding error based on 34 (33) place cells recorded during 10 min of foraging was 8.0 (7.7) cm. Our statistical paradigm provides a reliable approach for quantifying the spatial information in the ensemble place cell firing patterns and defines a generally applicable framework for studying information encoding in neural systems.
Figures
References
-
- Abbott LF. Decoding neuronal firing and modeling neural networks. Q Rev Biophys. 1994;27:291–331. - PubMed
-
- Bialek W, Zee A. Coding and computation with neural spike trains. J Stat Phys. 1990;59:103–115.
-
- Bialek W, Rieke F, de Ruyter van Steveninck RR, Warland D. Reading a neural code. Science. 1991;252:1854–1857. - PubMed
-
- Box GEP, Jenkins GM, Reinsel GC. Time series analysis: forecasting and control, Ed 3. Prentice Hall; Englewood Cliffs, NJ: 1994.
-
- Brown EN, Frank LM, Wilson MA. Statistical approaches to place field estimation and neuronal ensemble decoding. Soc Neurosci Abstr. 1996;22:910.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
