HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors
- PMID: 9736741
- PMCID: PMC21647
- DOI: 10.1073/pnas.95.19.11360
HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors
Abstract
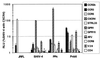
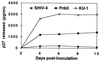
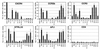
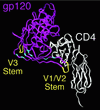
The envelope (Env) proteins of primate lentiviruses interact sequentially with CD4 and a coreceptor to infect cells. Changes in coreceptor use strongly influence viral tropism and pathogenesis. We followed the evolution of coreceptor use in pig-tailed macaques that developed severe CD4 T-cell loss during the derivation of a pathogenic simian HIV (SHIV) that contained the tat, rev, vpu, and env genes of the HXBc2 strain of HIV-1 in a genetic background of SIVmac239. The Env from the parental virus as well as one derived from the first macaque to develop AIDS exclusively used CXCR4 as a coreceptor, indicating that CXCR4 can function as a coreceptor in macaques even though it is rarely used by simian immunodeficiency viruses. One Env (Pnb5), obtained from a macrophage-tropic virus isolated from the cerebral spinal fluid, did not use CCR5 or CXCR4. Instead, it used CCR2b and to a lesser extent CCR3, STRL33, and APJ to infect cells. Chimeras between Pnb5 and the parental X4 Env indicated that the V3 loop is the major determinant of CXCR4 use, with other regions of Env influencing the efficiency with which this coreceptor was used. In contrast, the Pnb5 V1/2 and V3 regions in combination were both necessary and sufficient to confer full use of CCR2b, CCR3, STRL33, and APJ to the parental X4 Env protein. These results are consistent with a single, conserved binding site in Env that interacts with multiple coreceptors in conjunction with the V1/2 and V3 loops, and suggest that the V1/2 region plays a more important role in governing the use of CCR2b, CCR3, STRL33, and APJ than for CXCR4.
Figures

References
-
- Doms R W, Peiper S C. Virology. 1997;235:179–190. - PubMed
-
- Broder C C, Collman R G. J Leukocyte Biol. 1997;62:20–29. - PubMed
-
- Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. - PubMed
-
- Bieniasz P D, Cullen B R. Front Biosci. 1998;3:44–58. - PubMed
-
- Berger E A. AIDS. 1997;11:S3–S16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

