Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine
- PMID: 9736747
- PMCID: PMC21653
- DOI: 10.1073/pnas.95.19.11394
Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine
Abstract
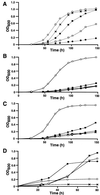
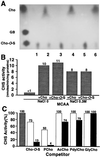
Glycine betaine is a potent osmoprotectant accumulated by Sinorhizobium meliloti to cope with osmotic stress. The biosynthesis of glycine betaine from choline is encoded by an operon of four genes, betICBA, as determined by sequence and mutant analysis. The betI and betC genes are separated by an intergenic region containing a 130-bp mosaic element that also is present between the betB and betA genes. In addition to the genes encoding a presumed regulatory protein (betI), the betaine aldehyde dehydrogenase (betB), and the choline dehydrogenase (betA) enzymes also found in Escherichia coli, a new gene (betC) was identified as encoding a choline sulfatase catalyzing the conversion of choline-O-sulfate and, at a lower rate, phosphorylcholine, into choline. Choline sulfatase activity was absent from betC but not from betB mutants and was shown to be induced indifferently by choline or choline-O-sulfate as were the other enzymes of the pathway. Unlike what has been shown in other bacteria and plants, choline-O-sulfate is not used as an osmoprotectant per se in S. meliloti, but is metabolized into glycine betaine. S. meliloti also can use this compound as the sole carbon, nitrogen, and sulfur source for growth and that depends on a functional bet locus. In conclusion, choline-O-sulfate and phosphorylcholine, which are found in higher plants and fungi, appear to be substrates for glycine betaine biosynthesis in S. meliloti.
Figures
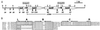

References
-
- Le Rudulier D, Strøm A R, Dandekar A M, Smith L T, Valentine R C. Science. 1984;224:1064–1068. - PubMed
-
- Csonka L N, Hanson A D. Annu Rev Microbiol. 1991;45:569–606. - PubMed
-
- Rhodes D, Hanson A D. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:357–384.
-
- Miller K J, Wood J M. Annu Rev Microbiol. 1996;50:101–136. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

